Expanding the scope of native chemical ligation - templated small molecule drug synthesis via benzanilide formation
- PMID: 34777764
- PMCID: PMC8528049
- DOI: 10.1039/d1sc00513h
Expanding the scope of native chemical ligation - templated small molecule drug synthesis via benzanilide formation
Abstract
We describe a reaction system that enables the synthesis of Bcr-Abl tyrosine kinase inhibitors (TKI) via benzanilide formation in water. The reaction is based on native chemical ligation (NCL). In contrast to previous applications, we used the NCL chemistry to establish aromatic rather than aliphatic amide bonds in coupling reactions between benzoyl and o-mercaptoaniline fragments. The method was applied for the synthesis of thiolated ponatinib and GZD824 derivatives. Acid treatment provided benzothiazole structures, which opens opportunities for diversification. Thiolation affected the affinity for Abl1 kinase only moderately. Of note, a ponatinib-derived benzothiazole also showed nanomolar affinity. NCL-enabled benzanilide formation may prove useful for fragment-based drug discovery. To show that benzanilide synthesis can be put under the control of a template, we connected the benzoyl and o-mercaptoaniline fragments to DNA and peptide nucleic acid (PNA) oligomers. Complementary RNA templates enabled adjacent binding of reactive conjugates triggering a rapid benzoyl transfer from a thioester-linked DNA conjugate to an o-mercaptoaniline-DNA or -PNA conjugate. We evaluated the influence of linker length and unpaired spacer nucleotides within the RNA template on the product yield. The data suggest that nucleic acid-templated benzanilide formation could find application in the establishment of DNA-encoded combinatorial libraries (DEL).
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures






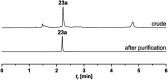


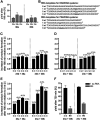
Similar articles
-
Templated native chemical ligation: peptide chemistry beyond protein synthesis.J Pept Sci. 2014 Feb;20(2):78-86. doi: 10.1002/psc.2602. Epub 2014 Jan 7. J Pept Sci. 2014. PMID: 24395765 Review.
-
RNA-templated chemical synthesis of proapoptotic L- and d-peptides.Bioorg Med Chem. 2022 Jul 15;66:116786. doi: 10.1016/j.bmc.2022.116786. Epub 2022 May 14. Bioorg Med Chem. 2022. PMID: 35594647
-
Reducing product inhibition in nucleic acid-templated ligation reactions: DNA-templated cycligation.Chembiochem. 2013 Nov 25;14(17):2322-8. doi: 10.1002/cbic.201300516. Epub 2013 Oct 9. Chembiochem. 2013. PMID: 24243697
-
Towards a templated reaction that translates RNA in cells into a proaptotic peptide-PNA conjugate.J Pept Sci. 2023 Jul;29(7):e3477. doi: 10.1002/psc.3477. Epub 2023 Jan 20. J Pept Sci. 2023. PMID: 36606596
-
Nucleic acids as templates and catalysts in chemical reactions: target-guided dynamic combinatorial chemistry and in situ click chemistry and DNA/RNA induced enantioselective reactions.Chem Soc Rev. 2023 Jul 3;52(13):4248-4291. doi: 10.1039/d3cs00166k. Chem Soc Rev. 2023. PMID: 37306487 Review.
References
-
- Liu L. Isr. J. Chem. 2019;59:64–70. doi: 10.1002/ijch.201800135. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

