Metagenomic Analysis of Dental Plaque on Pit and Fissure Sites With and Without Caries Among Adolescents
- PMID: 34778105
- PMCID: PMC8579706
- DOI: 10.3389/fcimb.2021.740981
Metagenomic Analysis of Dental Plaque on Pit and Fissure Sites With and Without Caries Among Adolescents
Abstract
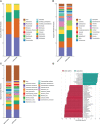
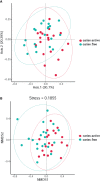
Caries is one of the most prevalent infectious diseases worldwide and is driven by the dysbiosis of dental biofilms adhering to tooth surfaces. The pits and fissured surfaces are the most susceptible sites of caries. However, information on the taxonomic composition and functional characteristics of the plaque microbiota in the pit and fissure sites is very limited. This study aimed to use metagenomic sequencing analyses to investigate the relationship between the plaque microbiome in the pit and fissure site and caries in adolescents. A total of 20 adolescents with active pit and fissure surface caries were involved as well as 20 age-matched, caries-free teenagers for control tests. Plaque samples were collected from the pit and fissure site and were subjected to metagenomic analyses, in which the microbial communities were investigated. Our results showed that the microbiota diversity was similar between those two groups. At the species level, the relative abundances of A. gerencseriae, P. acidifaciens, P. multisaccharivorax, S. oralis, S. mutans, and P. denticolens were higher in the caries-active group. N. elongata, C. hominis, and A. johnsonii were relatively more abundant in the caries-free groups. Functional analysis suggested that the metabolic pathway was the most abundant pathway, and the functional traits of the level 2 pathways included amino acid metabolism, metabolism of cofactors, and vitamins and carbohydrate metabolism. Our results also revealed that the caries group displayed several alterations in metabolic pathways, including enriched functions in carbohydrate digestion and absorption. This study suggested that in addition to the specific anatomical structures of the pit and fissured surfaces, the fundamental differences in the plaque microbiome may also be related to the susceptibility of pit and fissure caries.
Keywords: biofilms; bioinformatics; biomarkers; dental caries; microbiome.
Copyright © 2021 Pang, Wang, Ye, Zhou, Zhi and Lin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Dental plaque microbiota profiles of children with caries-free and caries-active dentition.J Dent. 2021 Jan;104:103539. doi: 10.1016/j.jdent.2020.103539. Epub 2020 Nov 25. J Dent. 2021. PMID: 33248211
-
Microbiomes of Site-Specific Dental Plaques from Children with Different Caries Status.Infect Immun. 2017 Jul 19;85(8):e00106-17. doi: 10.1128/IAI.00106-17. Print 2017 Aug. Infect Immun. 2017. PMID: 28507066 Free PMC article.
-
Protein relative abundance patterns associated with sucrose-induced dysbiosis are conserved across taxonomically diverse oral microcosm biofilm models of dental caries.Microbiome. 2015 Dec 19;3:69. doi: 10.1186/s40168-015-0136-z. Microbiome. 2015. PMID: 26684897 Free PMC article.
-
The Burden and Management of Dental Caries in Older Children.Pediatr Clin North Am. 2018 Oct;65(5):955-963. doi: 10.1016/j.pcl.2018.05.005. Pediatr Clin North Am. 2018. PMID: 30213356 Review.
-
Does assessment of microbial composition of plaque/saliva allow for diagnosis of disease activity of individuals?Community Dent Oral Epidemiol. 1997 Feb;25(1):76-81. doi: 10.1111/j.1600-0528.1997.tb00902.x. Community Dent Oral Epidemiol. 1997. PMID: 9088695 Review.
Cited by
-
How probiotics, prebiotics, synbiotics, and postbiotics prevent dental caries: an oral microbiota perspective.NPJ Biofilms Microbiomes. 2024 Feb 24;10(1):14. doi: 10.1038/s41522-024-00488-7. NPJ Biofilms Microbiomes. 2024. PMID: 38402294 Free PMC article. Review.
-
The Oral Microbiome: A Key Determinant of Oral Health.Adv Exp Med Biol. 2025;1472:133-149. doi: 10.1007/978-3-031-79146-8_9. Adv Exp Med Biol. 2025. PMID: 40111690 Review.
-
Elimination of Pathogen Biofilms via Postbiotics from Lactic Acid Bacteria: A Promising Method in Food and Biomedicine.Microorganisms. 2024 Mar 30;12(4):704. doi: 10.3390/microorganisms12040704. Microorganisms. 2024. PMID: 38674648 Free PMC article. Review.
-
Culturing the Human Oral Microbiota, Updating Methodologies and Cultivation Techniques.Microorganisms. 2023 Mar 24;11(4):836. doi: 10.3390/microorganisms11040836. Microorganisms. 2023. PMID: 37110259 Free PMC article. Review.
-
The microbiome alterations of supragingival plaque among adolescents using clear aligners: a metagenomic sequencing analysis.Prog Orthod. 2024 Dec 16;25(1):48. doi: 10.1186/s40510-024-00547-x. Prog Orthod. 2024. PMID: 39676101 Free PMC article.
References
-
- Colombo A. P., Boches S. K., Cotton S. L., Goodson J. M., Kent R., Haffajee A. D., et al. . (2009). Comparisons of Subgingival Microbial Profiles of Refractory Periodontitis, Severe Periodontitis, and Periodontal Health Using the Human Oral Microbe Identification Microarray. J. Periodontol. 80 (9), 1421–1432. doi: 10.1902/jop.2009.090185 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

