Molecular Epidemiology of Extraintestinal Pathogenic Escherichia coli Causing Hemorrhagic Pneumonia in Mink in Northern China
- PMID: 34778114
- PMCID: PMC8581539
- DOI: 10.3389/fcimb.2021.781068
Molecular Epidemiology of Extraintestinal Pathogenic Escherichia coli Causing Hemorrhagic Pneumonia in Mink in Northern China
Abstract
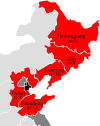
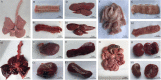
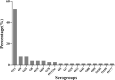
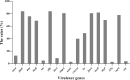
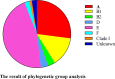
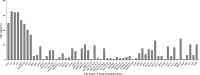
The molecular epidemiology and biological characteristics of Escherichia coli associated with hemorrhagic pneumonia (HP) mink from five Chinese Provinces were determined. From 2017 to 2019, 85 E. coli strains were identified from 115 lung samples of mink suffering from HP. These samples were subjected to serotyping, antimicrobial susceptibility, detection of virulence genes, phylogenetic grouping, whole-genome sequencing, drug resistant gene, multilocus sequence typing (MLST) and biofilm-forming assays. E. coli strains were divided into 18 serotypes. Thirty-nine E. coli strains belonged to the O11 serotype. Eighty-five E. coli strains were classified into seven phylogenetic groups: E (45.9%, 39/85), A (27.1%, 23/85), B1 (14.1%, 12/85), B2 (3.7%, 3/85), D (3.7%, 3/85), F (2.4%, 2/85) and clade I (1.2%, 1/85). MLST showed that the main sequence types (STs) were ST457 (27/66), All E. coli strains had ≥4 virulence genes. The prevalence of virulence was 98.8% for yijp and fimC, 96.5% for iucD, 95.3% for ompA, 91.8% for cnf-Ⅰ, 89.4% for mat, 82.3% for hlyF, and 81.2% for ibeB. The prevalence of virulence genes iss, cva/cvi, aatA, ibeA, vat, hlyF, and STa was 3.5-57.6%. All E. coli strains were sensitive to sulfamethoxazole, but high resistance was shown to tetracycline (76.5%), chloramphenicol (71.8%), ciprofloxacin (63.5%) and florfenicol (52.9%), resistance to other antibiotics was 35.3-16.5%. The types and ratios of drug-resistance genes were tet(A), strA, strB, sul2, oqxA, blaTEM-1B, floR, and catA1 had the highest frequency from 34%-65%, which were consistent with our drug resistance phenotype tetracycline, florfenicol, quinolones, chloramphenicol, the bla-NDM-I and mcr-I were presented in ST457 strains. Out of 85 E. coli strains, six (7.1%) possessed a strong ability, 12 (14.1%) possessed a moderate ability, and 64 (75.3%) showed a weak ability to form biofilm. Our data will aid understanding of the epidemiological background and provide a clinical basis for HP treatment in mink caused by E. coli.
Keywords: Escherichia coli; hemorrhagic pneumonia; multilocus sequence typing; serotype; virulence.
Copyright © 2021 Yu, Hu, Fan, Zhang, Lian, Li, Li, Yan, Wang and Bai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

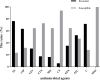

Similar articles
-
Genetic diversity and multidrug resistance of phylogenic groups B2 and D in InPEC and ExPEC isolated from chickens in Central China.BMC Microbiol. 2022 Feb 18;22(1):60. doi: 10.1186/s12866-022-02469-2. BMC Microbiol. 2022. PMID: 35180845 Free PMC article.
-
Antimicrobial Resistance and Virulence Profiles of mcr-1-Positive Escherichia coli Isolated from Swine Farms in Heilongjiang Province of China.J Food Prot. 2020 Dec 1;83(12):2209-2215. doi: 10.4315/JFP-20-190. J Food Prot. 2020. PMID: 32730609
-
Characterization and virulence clustering analysis of extraintestinal pathogenic Escherichia coli isolated from swine in China.BMC Vet Res. 2017 Apr 8;13(1):94. doi: 10.1186/s12917-017-0975-x. BMC Vet Res. 2017. PMID: 28388949 Free PMC article.
-
Bacterial characteristics of importance for recurrent urinary tract infections caused by Escherichia coli.Dan Med Bull. 2011 Apr;58(4):B4187. Dan Med Bull. 2011. PMID: 21466767 Review.
-
Molecular Epidemiology of Extraintestinal Pathogenic Escherichia coli.EcoSal Plus. 2018 Apr;8(1):10.1128/ecosalplus.ESP-0004-2017. doi: 10.1128/ecosalplus.ESP-0004-2017. EcoSal Plus. 2018. PMID: 29667573 Free PMC article. Review.
Cited by
-
A Novel CMY Variant Confers Transferable High-Level Resistance to Ceftazidime-Avibactam in Multidrug-Resistant Escherichia coli.Microbiol Spectr. 2023 Feb 14;11(2):e0334922. doi: 10.1128/spectrum.03349-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36786629 Free PMC article.
-
Antibiotic-Resistant Escherichia coli Strains Isolated from Captive Giant Pandas: A Reservoir of Antibiotic Resistance Genes and Virulence-Associated Genes.Vet Sci. 2022 Dec 18;9(12):705. doi: 10.3390/vetsci9120705. Vet Sci. 2022. PMID: 36548866 Free PMC article.
-
A virulent escherichia coli O121-B2-ST131 strain causes hemorrhagic pneumonia in mink: evidence from pathogenicity and animal challenge experiments.BMC Vet Res. 2025 May 27;21(1):378. doi: 10.1186/s12917-025-04817-6. BMC Vet Res. 2025. PMID: 40426189 Free PMC article.
-
Characterization of Extraintestinal Pathogenic Escherichia coli Strains Causing Canine Pneumonia in China: Antibiotic Resistance, Virulence Genes, and Sequence Typing.Vet Sci. 2024 Oct 10;11(10):491. doi: 10.3390/vetsci11100491. Vet Sci. 2024. PMID: 39453083 Free PMC article.
-
Emergence of NDM-producing Enterobacterales infections in companion animals from Argentina.BMC Vet Res. 2024 May 3;20(1):174. doi: 10.1186/s12917-024-04020-z. BMC Vet Res. 2024. PMID: 38702700 Free PMC article.
References
-
- Bancroft J. D., Stevens A. (1990). Theory and Practice of Histological Techniques, 3rd. Edinburgh: Churchill Livingstone. pp. 740.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

