A novel compound active against SARS-CoV-2 targeting uridylate-specific endoribonuclease (NendoU/NSP15): in silico and in vitro investigations
- PMID: 34778776
- PMCID: PMC8528204
- DOI: 10.1039/d1md00202c
A novel compound active against SARS-CoV-2 targeting uridylate-specific endoribonuclease (NendoU/NSP15): in silico and in vitro investigations
Abstract
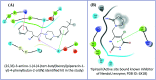
NendoU (NSP15) is an Mn(2+)-dependent, uridylate-specific enzyme, which leaves 2'-3'-cyclic phosphates 5' to the cleaved bond. Our in-house library was subjected to high throughput virtual screening (HTVS) to identify compounds with potential to inhibit NendoU enzyme, high-rank compounds (those that bound to multiple target structures) were further subjected to 100 nanoseconds MD simulations. Among these, one was found to be bound highly stable within the active site of the NendoU protein structure. Here, we are reporting a derivative of piperazine based '(2S,3S)-3-amino-1-(4-(4-(tert-butyl)benzyl)piperazin-1-yl)-4-phenylbutan-2-ol' (IV) from our in-house libraries having potential efficacy against SARS-CoV-2 in in vitro assays. This compound demonstrated inhibition of viral replication at the same level as Ivermectin, a known SARS-CoV-2 inhibitor, which is not used due to its toxicity at a higher than the currently approved dosage. Compound IV was not toxic to the cell lines up to a 50 μM concentration and exhibited IC50s of 4.97 μM and 8.46 μM in viral entry and spread assay, respectively. Therefore, this novel class of NendoU inhibitor could provide new insights for the development of treatment options for COVID-19.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- ICTV Code (https://talk.ictvonline.org/information/w/ictv-information/383/ictv-code), (accessed December 25, 2020)
-
- McIntosh K., Coronaviruses: A Comparative Review, in Current Topics in Microbiology and Immunology / Ergebnisse der Mikrobiologie und Immunitätsforschung, ed. W. Arberet al., Springer, Berlin, Heidelberg, 1974, vol. 63, 10.1007/978-3-642-65775-7_3 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

