Integrative Genomic Analysis of Pediatric Myeloid-Related Acute Leukemias Identifies Novel Subtypes and Prognostic Indicators
- PMID: 34778799
- PMCID: PMC8580615
- DOI: 10.1158/2643-3230.BCD-21-0049
Integrative Genomic Analysis of Pediatric Myeloid-Related Acute Leukemias Identifies Novel Subtypes and Prognostic Indicators
Abstract
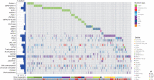
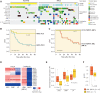
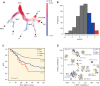
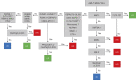
Genomic characterization of pediatric patients with acute myeloid leukemia (AML) has led to the discovery of somatic mutations with prognostic implications. Although gene-expression profiling can differentiate subsets of pediatric AML, its clinical utility in risk stratification remains limited. Here, we evaluate gene expression, pathogenic somatic mutations, and outcome in a cohort of 435 pediatric patients with a spectrum of pediatric myeloid-related acute leukemias for biological subtype discovery. This analysis revealed 63 patients with varying immunophenotypes that span a T-lineage and myeloid continuum designated as acute myeloid/T-lymphoblastic leukemia (AMTL). Within AMTL, two patient subgroups distinguished by FLT3-ITD and PRC2 mutations have different outcomes, demonstrating the impact of mutational composition on survival. Across the cohort, variability in outcomes of patients within isomutational subsets is influenced by transcriptional identity and the presence of a stem cell-like gene-expression signature. Integration of gene expression and somatic mutations leads to improved risk stratification.
Significance: Immunophenotype and somatic mutations play a significant role in treatment approach and risk stratification of acute leukemia. We conducted an integrated genomic analysis of pediatric myeloid malignancies and found that a combination of genetic and transcriptional readouts was superior to immunophenotype and genomic mutations in identifying biological subtypes and predicting outcomes. This article is highlighted in the In This Issue feature, p. 549.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures


References
-
- Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med 1985;103:620–5. - PubMed
-
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–405. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

