Machine learning and network medicine approaches for drug repositioning for COVID-19
- PMID: 34778851
- PMCID: PMC8576113
- DOI: 10.1016/j.patter.2021.100396
Machine learning and network medicine approaches for drug repositioning for COVID-19
Abstract
We present two machine learning approaches for drug repurposing. While we have developed them for COVID-19, they are disease-agnostic. The two methodologies are complementary, targeting SARS-CoV-2 and host factors, respectively. Our first approach consists of a matrix factorization algorithm to rank broad-spectrum antivirals. Our second approach, based on network medicine, uses graph kernels to rank drugs according to the perturbation they induce on a subnetwork of the human interactome that is crucial for SARS-CoV-2 infection/replication. Our experiments show that our top predicted broad-spectrum antivirals include drugs indicated for compassionate use in COVID-19 patients; and that the ranking obtained by our kernel-based approach aligns with experimental data. Finally, we present the COVID-19 repositioning explorer (CoREx), an interactive online tool to explore the interplay between drugs and SARS-CoV-2 host proteins in the context of biological networks, protein function, drug clinical use, and Connectivity Map. CoREx is freely available at: https://paccanarolab.org/corex/.
Keywords: COVID-19; SARS-CoV-2; drug repurposing; graph visualization; kernels on graphs; network medicine; non-negative matrix factorization.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures
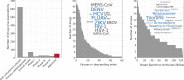

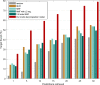

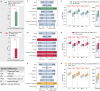

Comment in
-
AI in drug discovery: Applications, opportunities, and challenges.Patterns (N Y). 2022 Jun 10;3(6):100529. doi: 10.1016/j.patter.2022.100529. eCollection 2022 Jun 10. Patterns (N Y). 2022. PMID: 35755871 Free PMC article. No abstract available.
References
-
- Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3:673–683. - PubMed
-
- Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C., et al. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2018;18:41–58. - PubMed
-
- Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses —drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. https://www.nature.com/articles/nrd.2015.37 - DOI - PMC - PubMed
-
- Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

