The taxonomy, host range and pathogenicity of coronaviruses and other viruses in the Nidovirales order
- PMID: 34778878
- PMCID: PMC8062217
- DOI: 10.1186/s44149-021-00005-9
The taxonomy, host range and pathogenicity of coronaviruses and other viruses in the Nidovirales order
Abstract
The frequent emergence of coronavirus (CoV) epidemics has seriously threatened public health and stock farming. The major hosts for CoVs are birds and mammals. Although most CoVs inhabit their specific natural hosts, some may occasionally cross the host barrier to infect livestock and even people, causing a variety of diseases. Since the beginning of the new century, increasing attention has been given to research on CoVs due to the emergence of highly pathogenic and genetically diverse CoVs that have caused several epidemics, including the recent COVID-19 pandemic. CoVs belong to the Coronaviridae family of the Nidovirales order. Recently, advanced techniques for viral detection and viral genome analyses have enabled characterization of many new nidoviruses than ever and have greatly expanded the Nidovirales order with new classification and nomenclature. Here, we first provide an overview of the latest research progress in the classification of the Nidovirales order and then introduce the host range, genetic variation, genomic pattern and pathogenic features of epidemic CoVs and other epidemic viruses. This information will promote understanding of the phylogenetic relationship and infectious transmission of various pathogenic nidoviruses, including epidemic CoVs, which will benefit virological research and viral disease control.
Keywords: Coronavirus; Evolution; Genetics; Hosts; Nidovirales; S protein.
© The Author(s) 2021.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures


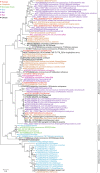
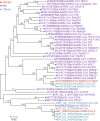
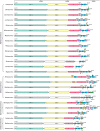

References
-
- Bailey AL, Lauck M, Weiler A, Sibley SD, Dinis JM, Bergman Z, Nelson CW, Correll M, Gleicher M, Hyeroba D, et al. High genetic diversity and adaptive potential of two simian hemorrhagic fever viruses in a wild primate population. PLoS One. 2014;9(3):e90714. doi: 10.1371/journal.pone.0090714. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
