In silico characterisation of the complete Ly6 protein family in Fasciola gigantica supported through transcriptomics of the newly-excysted juveniles
- PMID: 34781332
- PMCID: PMC8763315
- DOI: 10.1039/d1mo00254f
In silico characterisation of the complete Ly6 protein family in Fasciola gigantica supported through transcriptomics of the newly-excysted juveniles
Abstract
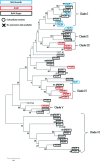
Fasciola gigantica is one of the aetiological trematodes associated with fascioliasis, which heavily impacts food-production systems and human and animal welfare on a global scale. In the absence of a vaccine, fascioliasis control and treatment is restricted to pasture management, such as clean grazing, and a limited array of chemotherapies, to which signs of resistance are beginning to appear. Research into novel control strategies is therefore urgently required and the advent of 'omics technologies presents considerable opportunity for novel drug and vaccine target discovery. Here, interrogation of the first available F. gigantica newly excysted juvenile (NEJ) transcriptome revealed several protein families of current interest to parasitic flatworm vaccine research, including orthologues of mammalian complement regulator CD59 of the Ly6 family. Ly6 proteins have previously been identified on the tegument of Schistosoma mansoni and induced protective immunity in vaccination trials. Incorporating the recently available F. gigantica genome, the current work revealed 20 novel Ly6 family members in F. gigantica and, in parallel, significantly extended the F. hepatica complement from 3 to 18 members. Phylogenetic analysis revealed several distinct clades within the family, some of which are unique to Fasciola spp. trematodes. Analysis of available proteomic databases also revealed three of the newly discovered FhLy6s were present in extracellular vesicles, which have previously been prioritised in studying the host-parasite interface. The presentation of this new transcriptomic resource, in addition to the Ly6 family proteins here identified, represents a wealth of opportunity for future vaccine research.
Conflict of interest statement
The authors declare there are no conflicts of interest.
Figures


Similar articles
-
First insight into CD59-like molecules of adult Fasciola hepatica.Exp Parasitol. 2014 Sep;144:57-64. doi: 10.1016/j.exppara.2014.06.012. Epub 2014 Jun 21. Exp Parasitol. 2014. PMID: 24955521
-
A novel ex vivo immunoproteomic approach characterising Fasciola hepatica tegumental antigens identified using immune antibody from resistant sheep.Int J Parasitol. 2017 Aug;47(9):555-567. doi: 10.1016/j.ijpara.2017.02.004. Epub 2017 Apr 26. Int J Parasitol. 2017. PMID: 28455238
-
Molecular characterisation and vaccine efficacy of two novel developmentally regulated surface tegument proteins of Fasciola hepatica.Vet Parasitol. 2020 Oct;286:109244. doi: 10.1016/j.vetpar.2020.109244. Epub 2020 Sep 12. Vet Parasitol. 2020. PMID: 32971381
-
Omics tools enabling vaccine discovery against fasciolosis.Trends Parasitol. 2022 Dec;38(12):1068-1079. doi: 10.1016/j.pt.2022.09.009. Epub 2022 Oct 18. Trends Parasitol. 2022. PMID: 36270885 Review.
-
Differential expression of microRNAs and tRNA fragments mediate the adaptation of the liver fluke Fasciola gigantica to its intermediate snail and definitive mammalian hosts.Int J Parasitol. 2021 Apr;51(5):405-414. doi: 10.1016/j.ijpara.2020.10.009. Epub 2021 Jan 26. Int J Parasitol. 2021. PMID: 33513403 Review.
Cited by
-
Detoxome Capacity of the Adult Rumen Fluke Calicophoron daubneyi Extends into Its Secreted Extracellular Vesicles.J Proteome Res. 2025 Feb 7;24(2):624-638. doi: 10.1021/acs.jproteome.4c00615. Epub 2025 Jan 19. J Proteome Res. 2025. PMID: 39829022 Free PMC article.
-
Discovery of long non-coding RNAs in the liver fluke, Fasciola hepatica.PLoS Negl Trop Dis. 2023 Sep 28;17(9):e0011663. doi: 10.1371/journal.pntd.0011663. eCollection 2023 Sep. PLoS Negl Trop Dis. 2023. PMID: 37769025 Free PMC article.
-
Orientational Preferences of GPI-Anchored Ly6/uPAR Proteins.Int J Mol Sci. 2022 Dec 20;24(1):11. doi: 10.3390/ijms24010011. Int J Mol Sci. 2022. PMID: 36613456 Free PMC article.
-
Proteomic analysis of Fasciola gigantica excretory and secretory products (FgESPs) co-immunoprecipitated using a time course of infected buffalo sera.Front Microbiol. 2022 Dec 23;13:1089394. doi: 10.3389/fmicb.2022.1089394. eCollection 2022. Front Microbiol. 2022. PMID: 36620027 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

