Evolutionary and Phenotypic Characterization of Two Spike Mutations in European Lineage 20E of SARS-CoV-2
- PMID: 34781748
- PMCID: PMC8593680
- DOI: 10.1128/mBio.02315-21
Evolutionary and Phenotypic Characterization of Two Spike Mutations in European Lineage 20E of SARS-CoV-2
Abstract
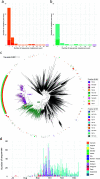
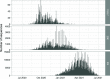
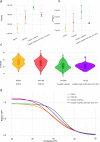
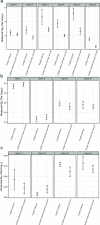
We have detected two mutations in the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at amino acid positions 1163 and 1167 that appeared independently in multiple transmission clusters and different genetic backgrounds. Furthermore, both mutations appeared together in a cluster of 1,627 sequences belonging to clade 20E. This cluster is characterized by 12 additional single nucleotide polymorphisms but no deletions. The available structural information on the S protein in the pre- and postfusion conformations predicts that both mutations confer rigidity, which could potentially decrease viral fitness. Accordingly, we observed reduced infectivity of this spike genotype relative to the ancestral 20E sequence in vitro, and the levels of viral RNA in nasopharyngeal swabs were not significantly higher. Furthermore, the mutations did not impact thermal stability or antibody neutralization by sera from vaccinated individuals but moderately reduce neutralization by convalescent-phase sera from the early stages of the pandemic. Despite multiple successful appearances of the two spike mutations during the first year of SARS-CoV-2 evolution, the genotype with both mutations was displaced upon the expansion of the 20I (Alpha) variant. The midterm fate of the genotype investigated was consistent with the lack of advantage observed in the clinical and experimental data. IMPORTANCE We observed repeated, independent emergence of mutations in the SARS-CoV-2 spike involving amino acids 1163 and 1167, within the HR2 functional motif. Conclusions derived from evolutionary and genomic diversity analysis suggest that the co-occurrence of both mutations might pose an advantage for the virus and therefore a threat to effective control of the epidemic. However, biological characterization, including in vitro experiments and analysis of clinical data, indicated no clear benefit in terms of stability or infectivity. In agreement with this, continuous epidemiological surveillance conducted months after the first observations revealed that both mutations did not successfully outcompete other variants and stopped circulating 9 months after their initial detection. Additionally, we evaluated the potential of both mutations to escape neutralizing antibodies, finding that the presence of these two mutations on their own is not likely to confer antibody escape. Our results provide an example of how newly emerged spike mutations can be assessed to better understand the risk posed by new variants and indicate that some spike mutations confer no clear advantage to the virus despite independently emerging multiple times and are eventually displaced by fitter variants.
Keywords: HR2; SARS-CoV-2; adaptive mutations; antibody escape; homoplasy; spike; variants.
Figures

References
-
- Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O'Toole Á, Southgate J, Johnson R, Jackson B, Nascimento FF, Rey SM, Nicholls SM, Colquhoun RM, da Silva Filipe A, Shepherd J, Pascall DJ, Shah R, Jesudason N, Li K, Jarrett R, Pacchiarini N, Bull M, Geidelberg L, Siveroni I, Goodfellow I, Loman NJ, Pybus OG, Robertson DL, Thomson EC, Rambaut A, Connor TR, COG-UK Consortium. 2021. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184:64–75.E11. doi:10.1016/j.cell.2020.11.020. - DOI - PMC - PubMed
-
- Hodcroft EB, Zuber M, Nadeau S, Vaughan TG, Crawford KHD, Althaus CL, Reichmuth ML, Bowen JE, Walls AC, Corti D, Bloom JD, Veesler D, Mateo D, Hernando A, Comas I, González-Candelas F, González-Candelas F, Goig GA, Chiner-Oms Á, Cancino-Muñoz I, López MG, Torres-Puente M, Gomez-Navarro I, Jiménez-Serrano S, Ruiz-Roldán L, Bracho MA, García-González N, Martínez-Priego L, Galán-Vendrell I, Ruiz-Hueso P, De Marco G, Ferrús ML, Carbó-Ramírez S, D’Auria G, Coscollá M, Ruiz-Rodríguez P, Roig-Sena FJ, Sanmartín I, Garcia-Souto D, Pequeno-Valtierra A, Tubio JMC, Rodríguez-Castro J, Rabella N, Navarro F, Miró E, Rodríguez-Iglesias M, Galán-Sanchez F, Rodriguez-Pallares S, de Toro M, Escudero MB, SeqCOVID-SPAIN consortium, et al. . 2021. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature 595:707–712. doi:10.1038/s41586-021-03677-y. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- European Commission-NextGenerationEU
- RTI2018-094399-A-I00/Ministerio de Ciencia, Innovación y Universidades (MCIU)
- COV20/00140/MEC | Instituto de Salud Carlos III (ISCIII)
- SEJI/2019/011/GVA | Conselleria de Innovación, Universidades, Ciencia y Sociedad Digital, Generalitat Valenciana (CIUCSD)
- CSIC-COV19-021/MEC | Consejo Superior de Investigaciones Científicas (CSIC)
LinkOut - more resources
Full Text Sources
Miscellaneous

