Fusobacterium nucleatum drives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression
- PMID: 34781821
- PMCID: PMC8604392
- DOI: 10.1080/19490976.2021.1987780
Fusobacterium nucleatum drives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression
Abstract
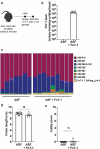
The colorectal cancer (CRC)-associated microbiota creates a pro-tumorigenic intestinal milieu and shapes immune responses within the tumor microenvironment. However, how oncomicrobes - like Fusobacterium nucleatum, found in the oral cavity and associated with CRC tissues- affect these distinct aspects of tumorigenesis is difficult to parse. Herein, we found that neonatal inoculation of ApcMin/+ mice with F. nucleatum strain Fn7-1 circumvents technical barriers preventing its intestinal colonization, drives colonic Il17a expression prior to tumor formation, and potentiates intestinal tumorigenesis. Using gnotobiotic mice colonized with a minimal complexity microbiota (the altered Schaedler's flora), we observed that intestinal Fn7-1 colonization increases colonic Th17 cell frequency and their IL-17A and IL-17F expression, along with a concurrent increase in colonic lamina propria Il23p19 expression. As Fn7-1 stably colonizes the intestinal tract in our models, we posited that microbial metabolites, specifically short-chain fatty acids (SCFA) that F. nucleatum abundantly produces in culture and, as we demonstrate, in the intestinal tract, might mediate part of its immunomodulatory effects in vivo. Supporting this hypothesis, we found that Fn7-1 did not alter RORγt+ CD4+T cell frequency in the absence of the SCFA receptor FFAR2. Taken together, our work suggests that F. nucleatum influences intestinal immunity by shaping Th17 responses in an FFAR2-dependent manner, although further studies are necessary to clarify the precise and multifaceted roles of FFAR2. The potential to increase intestinal Th17 responses is shared by another oncomicrobe, enterotoxigenic Bacteroides fragilis, highlighting a conserved pathway that could potentially be targeted to slow oncomicrobe-mediated CRC.
Keywords: Fusobacterium nucleatum; Microbiome; Th17 cells; altered Schaedler’s flora; colorectal cancer; gnotobiotics; interleukin 17.
Conflict of interest statement
W.S.G. is on the Scientific Advisory Boards of Senda Biosciences, Evelo Biosciences, SanaRx and Tenza Inc. and has consulted for BioMx, Empress Biosciences, GSK, X-Biotix, Janssen, and Merck.
Figures




References
-
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–19. doi:10.1016/j.chom.2013.07.007. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
