CRISPR and KRAS: a match yet to be made
- PMID: 34781949
- PMCID: PMC8591907
- DOI: 10.1186/s12929-021-00772-0
CRISPR and KRAS: a match yet to be made
Abstract
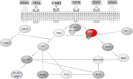
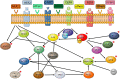
CRISPR (clustered regularly interspaced short palindromic repeats) systems are one of the most fascinating tools of the current era in molecular biotechnology. With the ease that they provide in genome editing, CRISPR systems generate broad opportunities for targeting mutations. Specifically in recent years, disease-causing mutations targeted by the CRISPR systems have been of main research interest; particularly for those diseases where there is no current cure, including cancer. KRAS mutations remain untargetable in cancer. Mutations in this oncogene are main drivers in common cancers, including lung, colorectal and pancreatic cancers, which are severe causes of public health burden and mortality worldwide, with no cure at hand. CRISPR systems provide an opportunity for targeting cancer causing mutations. In this review, we highlight the work published on CRISPR applications targeting KRAS mutations directly, as well as CRISPR applications targeting mutations in KRAS-related molecules. In specific, we focus on lung, colorectal and pancreatic cancers. To date, the limited literature on CRISPR applications targeting KRAS, reflect promising results. Namely, direct targeting of mutant KRAS variants using various CRISPR systems resulted in significant decrease in cell viability and proliferation in vitro, as well as tumor growth inhibition in vivo. In addition, the effect of mutant KRAS knockdown, via CRISPR, has been observed to exert regulatory effects on the downstream molecules including PI3K, ERK, Akt, Stat3, and c-myc. Molecules in the KRAS pathway have been subjected to CRISPR applications more often than KRAS itself. The aim of using CRISPR systems in these studies was mainly to analyze the therapeutic potential of possible downstream and upstream effectors of KRAS, as well as to discover further potential molecules. Although there have been molecules identified to have such potential in treatment of KRAS-driven cancers, a substantial amount of effort is still needed to establish treatment strategies based on these discoveries. We conclude that, at this point in time, despite being such a powerful directed genome editing tool, CRISPR remains to be underutilized for targeting KRAS mutations in cancer. Efforts channelled in this direction, might pave the way in solving the long-standing challenge of targeting the KRAS mutations in cancers.
Keywords: CRISPR; CRISPR-Cas; Cancer; Colon cancer; Genome editing; KRAS; Lung cancer; Mutation; NSCLC; Pancreatic cancer.
© 2021. The Author(s).
Conflict of interest statement
Authors declare no competing interests.
Figures
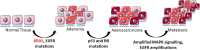


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

