Alternative splicing of NF-YA promotes prostate cancer aggressiveness and represents a new molecular marker for clinical stratification of patients
- PMID: 34782004
- PMCID: PMC8594157
- DOI: 10.1186/s13046-021-02166-4
Alternative splicing of NF-YA promotes prostate cancer aggressiveness and represents a new molecular marker for clinical stratification of patients
Abstract
Background: Approaches based on expression signatures of prostate cancer (PCa) have been proposed to predict patient outcomes and response to treatments. The transcription factor NF-Y participates to the progression from benign epithelium to both localized and metastatic PCa and is associated with aggressive transcriptional profile. The gene encoding for NF-YA, the DNA-binding subunit of NF-Y, produces two alternatively spliced transcripts, NF-YAs and NF-YAl. Bioinformatic analyses pointed at NF-YA splicing as a key transcriptional signature to discriminate between different tumor molecular subtypes. In this study, we aimed to determine the pathophysiological role of NF-YA splice variants in PCa and their association with aggressive subtypes.
Methods: Data on the expression of NF-YA isoforms were extracted from the TCGA (The Cancer Genome Atlas) database of tumor prostate tissues and validated in prostate cell lines. Lentiviral transduction and CRISPR-Cas9 technology allowed the modulation of the expression of NF-YA splice variants in PCa cells. We characterized 3D cell cultures through in vitro assays and RNA-seq profilings. We used the rank-rank hypergeometric overlap approach to identify concordant/discordant gene expression signatures of NF-YAs/NF-YAl-overexpressing cells and human PCa patients. We performed in vivo studies in SHO-SCID mice to determine pathological and molecular phenotypes of NF-YAs/NF-YAl xenograft tumors.
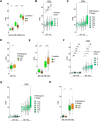
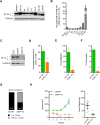
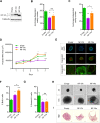
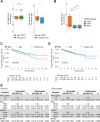
Results: NF-YA depletion affects the tumorigenic potential of PCa cells in vitro and in vivo. Elevated NF-YAs levels are associated to aggressive PCa specimens, defined by Gleason Score and TNM classification. NF-YAl overexpression increases cell motility, while NF-YAs enhances cell proliferation in PCa 3D spheroids and xenograft tumors. The transcriptome of NF-YAs-spheroids has an extensive overlap with localized and metastatic human PCa signatures. According to PCa PAM50 classification, NF-YAs transcript levels are higher in LumB, characterized by poor prognosis compared to LumA and basal subtypes. A significant decrease in NF-YAs/NF-YAl ratio distinguishes PCa circulating tumor cells from cancer cells in metastatic sites, consistently with pro-migratory function of NF-YAl. Stratification of patients based on NF-YAs expression is predictive of clinical outcome.
Conclusions: Altogether, our results indicate that the modulation of NF-YA isoforms affects prostate pathophysiological processes and contributes to cancer-relevant phenotype, in vitro and in vivo. Evaluation of NF-YA splicing may represent a new molecular strategy for risk assessment of PCa patients.
Keywords: Alternative splicing; Genome editing; NF-Y; Prostate cancer; Transcriptome profiling.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
NF-YA Overexpression in Lung Cancer: LUSC.Genes (Basel). 2019 Nov 17;10(11):937. doi: 10.3390/genes10110937. Genes (Basel). 2019. PMID: 31744190 Free PMC article.
-
NF-YA splice variants have different roles on muscle differentiation.Biochim Biophys Acta. 2016 Apr;1859(4):627-38. doi: 10.1016/j.bbagrm.2016.02.011. Epub 2016 Feb 26. Biochim Biophys Acta. 2016. PMID: 26921500
-
NF-YA Overexpression in Lung Cancer: LUAD.Genes (Basel). 2020 Feb 14;11(2):198. doi: 10.3390/genes11020198. Genes (Basel). 2020. PMID: 32075093 Free PMC article.
-
Expression and function of NF-Y subunits in cancer.Biochim Biophys Acta Rev Cancer. 2024 Mar;1879(2):189082. doi: 10.1016/j.bbcan.2024.189082. Epub 2024 Feb 1. Biochim Biophys Acta Rev Cancer. 2024. PMID: 38309445 Review.
-
Alternative splicing and biological heterogeneity in prostate cancer.Nat Rev Urol. 2009 Aug;6(8):454-60. doi: 10.1038/nrurol.2009.125. Nat Rev Urol. 2009. PMID: 19657379 Review.
Cited by
-
NFYAv1 is a Tumor-Promoting Transcript Associated with Poor Prognosis of Hepatocellular Carcinoma.Med Sci Monit. 2023 Jan 21;29:e938410. doi: 10.12659/MSM.938410. Med Sci Monit. 2023. PMID: 36680333 Free PMC article.
-
NFYA promotes malignant behavior of triple-negative breast cancer in mice through the regulation of lipid metabolism.Commun Biol. 2023 Jun 2;6(1):596. doi: 10.1038/s42003-023-04987-9. Commun Biol. 2023. PMID: 37268670 Free PMC article.
-
NFYA-mediated promotion of castration-resistant prostate cancer progression through EGR4 regulation.Discov Oncol. 2024 Oct 5;15(1):528. doi: 10.1007/s12672-024-01392-4. Discov Oncol. 2024. PMID: 39367986 Free PMC article.
-
Comprehensive analysis of prognosis-related alternative splicing events in ovarian cancer.RNA Biol. 2022 Jan;19(1):1007-1018. doi: 10.1080/15476286.2022.2113148. RNA Biol. 2022. PMID: 35980273 Free PMC article.
-
Identification of alternative splicing associated with clinical features: from pan-cancers to genitourinary tumors.Front Oncol. 2023 Sep 25;13:1249932. doi: 10.3389/fonc.2023.1249932. eCollection 2023. Front Oncol. 2023. PMID: 37810965 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

