CyFi-MAP: an interactive pathway-based resource for cystic fibrosis
- PMID: 34782688
- PMCID: PMC8592983
- DOI: 10.1038/s41598-021-01618-3
CyFi-MAP: an interactive pathway-based resource for cystic fibrosis
Abstract
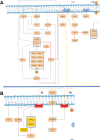
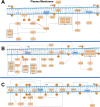
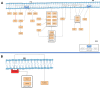
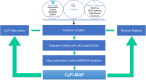
Cystic fibrosis (CF) is a life-threatening autosomal recessive disease caused by more than 2100 mutations in the CF transmembrane conductance regulator (CFTR) gene, generating variability in disease severity among individuals with CF sharing the same CFTR genotype. Systems biology can assist in the collection and visualization of CF data to extract additional biological significance and find novel therapeutic targets. Here, we present the CyFi-MAP-a disease map repository of CFTR molecular mechanisms and pathways involved in CF. Specifically, we represented the wild-type (wt-CFTR) and the F508del associated processes (F508del-CFTR) in separate submaps, with pathways related to protein biosynthesis, endoplasmic reticulum retention, export, activation/inactivation of channel function, and recycling/degradation after endocytosis. CyFi-MAP is an open-access resource with specific, curated and continuously updated information on CFTR-related pathways available online at https://cysticfibrosismap.github.io/ . This tool was developed as a reference CF pathway data repository to be continuously updated and used worldwide in CF research.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Saqi, M. et al. Medicine: The future of medical genomics, healthcare, and wellness. In Methods in Molecular Biology (2016). 10.1007/978-1-4939-3283-2_3 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

