Emerging technologies and their impact on regulatory science
- PMID: 34783606
- PMCID: PMC8749227
- DOI: 10.1177/15353702211052280
Emerging technologies and their impact on regulatory science
Abstract
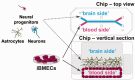
There is an evolution and increasing need for the utilization of emerging cellular, molecular and in silico technologies and novel approaches for safety assessment of food, drugs, and personal care products. Convergence of these emerging technologies is also enabling rapid advances and approaches that may impact regulatory decisions and approvals. Although the development of emerging technologies may allow rapid advances in regulatory decision making, there is concern that these new technologies have not been thoroughly evaluated to determine if they are ready for regulatory application, singularly or in combinations. The magnitude of these combined technical advances may outpace the ability to assess fit for purpose and to allow routine application of these new methods for regulatory purposes. There is a need to develop strategies to evaluate the new technologies to determine which ones are ready for regulatory use. The opportunity to apply these potentially faster, more accurate, and cost-effective approaches remains an important goal to facilitate their incorporation into regulatory use. However, without a clear strategy to evaluate emerging technologies rapidly and appropriately, the value of these efforts may go unrecognized or may take longer. It is important for the regulatory science field to keep up with the research in these technically advanced areas and to understand the science behind these new approaches. The regulatory field must understand the critical quality attributes of these novel approaches and learn from each other's experience so that workforces can be trained to prepare for emerging global regulatory challenges. Moreover, it is essential that the regulatory community must work with the technology developers to harness collective capabilities towards developing a strategy for evaluation of these new and novel assessment tools.
Keywords: Emerging technologies; bioimaging; bioinformatics; biomarkers; regulatory science; risk assessment.
Conflict of interest statement
Tim McCarthy is a shareholder of Pfizer.
Figures
































References
-
- Yamamoto E, Taquahashi Y, Kuwagata M, Saito H, Matsushita K, Toyoda T, Sato F, Kitajima S, Ogawa K, Izutsu K-i, Saito Y, Hirabayashi Y, Iimura Y, Honma M, Okuda H, Goda Y. Visualizing the spatial localization of ciclesonide and its metabolites in rat lungs after inhalation of 1-μm aerosol of ciclesonide by desorption electrospray ionization-time of flight mass spectrometry imaging. Int J Pharma 2021; 595:120241 - PubMed
-
- Lambert D, Pightling A, Griffiths E, Van Domselaar G, Evans P, Berthelet S, Craig D, Chandry PS, Stones R, Brinkman F, Angers-Loustau A, Kreysa J, Tong W, Blais B. Baseline practices for the application of genomic data supporting regulatory food safety. J AOAC Int 2017; 100:721–31 - PubMed
-
- Blais BW, Tapp K, Dixon M, Carrillo CD. Genomically informed strain-specific recovery of Shiga toxin-producing Escherichia coli during foodborne illness outbreak investigations. J Food Prot 2019; 82:39–44 - PubMed
Publication types
MeSH terms
Grants and funding
- UG3 TR003264/TR/NCATS NIH HHS/United States
- U2C ES030857/ES/NIEHS NIH HHS/United States
- R01 CA163256/CA/NCI NIH HHS/United States
- U24 TR002633/TR/NCATS NIH HHS/United States
- U2C ES030170/ES/NIEHS NIH HHS/United States
- U24 DK097193/DK/NIDDK NIH HHS/United States
- 001/WHO_/World Health Organization/International
- UG3 NS105703/NS/NINDS NIH HHS/United States
- P30 ES029067/ES/NIEHS NIH HHS/United States
- C45982/A21808/CRUK_/Cancer Research UK/United Kingdom
- U24 TR001950/TR/NCATS NIH HHS/United States
- P42 ES027704/ES/NIEHS NIH HHS/United States
- UH3 NS105703/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources

