Efferocytosis induces macrophage proliferation to help resolve tissue injury
- PMID: 34784501
- PMCID: PMC8665147
- DOI: 10.1016/j.cmet.2021.10.015
Efferocytosis induces macrophage proliferation to help resolve tissue injury
Abstract
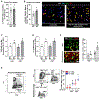
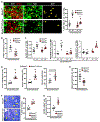
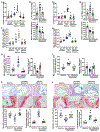
Apoptotic cell clearance by macrophages (efferocytosis) promotes resolution signaling pathways, which can be triggered by molecules derived from the phagolysosomal degradation of apoptotic cells. We show here that nucleotides derived from the hydrolysis of apoptotic cell DNA by phagolysosomal DNase2a activate a DNA-PKcs-mTORC2/Rictor pathway that increases Myc to promote non-inflammatory macrophage proliferation. Efferocytosis-induced proliferation expands the pool of resolving macrophages in vitro and in mice, including zymosan-induced peritonitis, dexamethasone-induced thymocyte apoptosis, and atherosclerosis regression. In the dexamethasone-thymus model, hematopoietic Rictor deletion blocked efferocytosing macrophage proliferation, apoptotic cell clearance, and tissue resolution. In atherosclerosis regression, silencing macrophage Rictor or DNase2a blocked efferocyte proliferation, apoptotic cell clearance, and plaque stabilization. In view of previous work showing that other types of apoptotic cell cargo can promote resolution in individual efferocytosing macrophages, the findings here suggest that signaling-triggered apoptotic cell-derived nucleotides can amplify this benefit by increasing the number of these macrophages.
Keywords: DNase2a; Erk1/2 signaling; MerTK; Myc; atherosclerosis; efferocytosis; inflammation resolution; mTORC2/Rictor; macrophage; macrophage proliferation.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Similar articles
-
Apoptosis recognition receptors regulate skin tissue repair in mice.Elife. 2023 Dec 21;12:e86269. doi: 10.7554/eLife.86269. Elife. 2023. PMID: 38127424 Free PMC article.
-
Myeloid MAS-driven macrophage efferocytosis promotes resolution in ischemia-stressed mouse and human livers.Sci Transl Med. 2025 Jul 9;17(806):eadr2725. doi: 10.1126/scitranslmed.adr2725. Epub 2025 Jul 9. Sci Transl Med. 2025. PMID: 40632838
-
Extracellular vesicles containing MFGE8 from colorectal cancer facilitate macrophage efferocytosis.Cell Commun Signal. 2024 May 27;22(1):295. doi: 10.1186/s12964-024-01669-9. Cell Commun Signal. 2024. PMID: 38802814 Free PMC article.
-
Anti-Atherogenic Mechanisms and Therapies.Curr Atheroscler Rep. 2025 Aug 20;27(1):83. doi: 10.1007/s11883-025-01324-9. Curr Atheroscler Rep. 2025. PMID: 40833492 Free PMC article. Review.
-
The role of efferocytosis-fueled macrophage metabolism in the resolution of inflammation.Immunol Rev. 2023 Oct;319(1):65-80. doi: 10.1111/imr.13214. Epub 2023 May 9. Immunol Rev. 2023. PMID: 37158427 Free PMC article. Review.
Cited by
-
Cross-comparison of systemic and tissue-specific metabolomes in a mouse model of Leigh syndrome.Metabolomics. 2021 Nov 18;17(12):101. doi: 10.1007/s11306-021-01854-8. Metabolomics. 2021. PMID: 34792662
-
The TRIM28/miR133a/CD47 axis acts as a potential therapeutic target in pancreatic necrosis by impairing efferocytosis.Mol Ther. 2024 Sep 4;32(9):3025-3041. doi: 10.1016/j.ymthe.2024.06.005. Epub 2024 Jun 12. Mol Ther. 2024. PMID: 38872307
-
The Role of Macrophage Efferocytosis in the Pathogenesis of Apical Periodontitis.Int J Mol Sci. 2024 Mar 29;25(7):3854. doi: 10.3390/ijms25073854. Int J Mol Sci. 2024. PMID: 38612664 Free PMC article.
-
In Situ Remodeling of Efferocytosis via Lesion-Localized Microspheres to Reverse Cartilage Senescence.Adv Sci (Weinh). 2024 May;11(19):e2400345. doi: 10.1002/advs.202400345. Epub 2024 Mar 13. Adv Sci (Weinh). 2024. PMID: 38477444 Free PMC article.
-
Efferocytosis drives a tryptophan metabolism pathway in macrophages to promote tissue resolution.Nat Metab. 2024 Sep;6(9):1736-1755. doi: 10.1038/s42255-024-01115-7. Epub 2024 Sep 6. Nat Metab. 2024. PMID: 39242914 Free PMC article.
References
-
- Alliot F, Godin I, and Pessac B (1999). Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res 117, 145–152. - PubMed
Publication types
MeSH terms
Grants and funding
- S10 RR027050/RR/NCRR NIH HHS/United States
- R35 HL145228/HL/NHLBI NIH HHS/United States
- S10 RR025686/RR/NCRR NIH HHS/United States
- R01 HL127464/HL/NHLBI NIH HHS/United States
- P30 CA013696/CA/NCI NIH HHS/United States
- P01 HL087123/HL/NHLBI NIH HHS/United States
- UL1 TR001873/TR/NCATS NIH HHS/United States
- R01 HL159012/HL/NHLBI NIH HHS/United States
- K99 HL145131/HL/NHLBI NIH HHS/United States
- S10 OD020056/OD/NIH HHS/United States
- T32 HL007343/HL/NHLBI NIH HHS/United States
- P20 GM121307/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

