Predicting the zoonotic capacity of mammals to transmit SARS-CoV-2
- PMID: 34784766
- PMCID: PMC8596006
- DOI: 10.1098/rspb.2021.1651
Predicting the zoonotic capacity of mammals to transmit SARS-CoV-2
Abstract
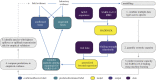
Back and forth transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) between humans and animals will establish wild reservoirs of virus that endanger long-term efforts to control COVID-19 in people and to protect vulnerable animal populations. Better targeting surveillance and laboratory experiments to validate zoonotic potential requires predicting high-risk host species. A major bottleneck to this effort is the few species with available sequences for angiotensin-converting enzyme 2 receptor, a key receptor required for viral cell entry. We overcome this bottleneck by combining species' ecological and biological traits with three-dimensional modelling of host-virus protein-protein interactions using machine learning. This approach enables predictions about the zoonotic capacity of SARS-CoV-2 for greater than 5000 mammals-an order of magnitude more species than previously possible. Our predictions are strongly corroborated by in vivo studies. The predicted zoonotic capacity and proximity to humans suggest enhanced transmission risk from several common mammals, and priority areas of geographic overlap between these species and global COVID-19 hotspots. With molecular data available for only a small fraction of potential animal hosts, linking data across biological scales offers a conceptual advance that may expand our predictive modelling capacity for zoonotic viruses with similarly unknown host ranges.
Keywords: COVID-19; ecological traits; machine learning; spillback; structural modelling; zoonotic.
Figures

Update of
-
Predicting the zoonotic capacity of mammals to transmit SARS-CoV-2.bioRxiv [Preprint]. 2021 Jun 29:2021.02.18.431844. doi: 10.1101/2021.02.18.431844. bioRxiv. 2021. Update in: Proc Biol Sci. 2021 Nov 24;288(1963):20211651. doi: 10.1098/rspb.2021.1651. PMID: 33619481 Free PMC article. Updated. Preprint.
Comment in
-
SARS-CoV-2 susceptibility of domestic and wild mammals: is it all about the similarity of their ACE2 receptor to the human one?Proc Biol Sci. 2022 Feb 9;289(1968):20212560. doi: 10.1098/rspb.2021.2560. Epub 2022 Feb 2. Proc Biol Sci. 2022. PMID: 35105235 Free PMC article. No abstract available.
References
-
- World Health Organization. 2021. WHO coronavirus disease (COVID-19) dashboard. See https://covid19.who.int.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
