Toward modeling metabolic state from single-cell transcriptomics
- PMID: 34785394
- PMCID: PMC8829761
- DOI: 10.1016/j.molmet.2021.101396
Toward modeling metabolic state from single-cell transcriptomics
Abstract
Background: Single-cell metabolic studies bring new insights into cellular function, which can often not be captured on other omics layers. Metabolic information has wide applicability, such as for the study of cellular heterogeneity or for the understanding of drug mechanisms and biomarker development. However, metabolic measurements on single-cell level are limited by insufficient scalability and sensitivity, as well as resource intensiveness, and are currently not possible in parallel with measuring transcript state, commonly used to identify cell types. Nevertheless, because omics layers are strongly intertwined, it is possible to make metabolic predictions based on measured data of more easily measurable omics layers together with prior metabolic network knowledge.
Scope of review: We summarize the current state of single-cell metabolic measurement and modeling approaches, motivating the use of computational techniques. We review three main classes of computational methods used for prediction of single-cell metabolism: pathway-level analysis, constraint-based modeling, and kinetic modeling. We describe the unique challenges arising when transitioning from bulk to single-cell modeling. Finally, we propose potential model extensions and computational methods that could be leveraged to achieve these goals.
Major conclusions: Single-cell metabolic modeling is a rising field that provides a new perspective for understanding cellular functions. The presented modeling approaches vary in terms of input requirements and assumptions, scalability, modeled metabolic layers, and newly gained insights. We believe that the use of prior metabolic knowledge will lead to more robust predictions and will pave the way for mechanistic and interpretable machine-learning models.
Keywords: Constraint-based modeling; Kinetic modeling; Metabolic modeling; Pathway analysis; Single-cell RNA-seq.
Copyright © 2021. Published by Elsevier GmbH.
Figures

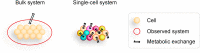


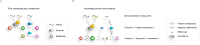
References
-
- Zhu C., Preissl S., Ren B. Single-cell multimodal omics: the power of many. Nature Methods. 2020;17(1):11–14. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

