Mediator dynamics during heat shock in budding yeast
- PMID: 34785526
- PMCID: PMC8744673
- DOI: 10.1101/gr.275750.121
Mediator dynamics during heat shock in budding yeast
Abstract
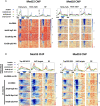
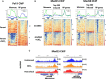
The Mediator complex is central to transcription by RNA polymerase II (Pol II) in eukaryotes. In budding yeast (Saccharomyces cerevisiae), Mediator is recruited by activators and associates with core promoter regions, where it facilitates preinitiation complex (PIC) assembly, only transiently before Pol II escape. Interruption of the transcription cycle by inactivation or depletion of Kin28 inhibits Pol II escape and stabilizes this association. However, Mediator occupancy and dynamics have not been examined on a genome-wide scale in yeast grown in nonstandard conditions. Here we investigate Mediator occupancy following heat shock or CdCl2 exposure, with and without depletion of Kin28. We find that Pol II occupancy shows similar dependence on Mediator under normal and heat shock conditions. However, although Mediator association increases at many genes upon Kin28 depletion under standard growth conditions, little or no increase is observed at most genes upon heat shock, indicating a more stable association of Mediator after heat shock. Unexpectedly, Mediator remains associated upstream of the core promoter at genes repressed by heat shock or CdCl2 exposure whether or not Kin28 is depleted, suggesting that Mediator is recruited by activators but is unable to engage PIC components at these repressed targets. This persistent association is strongest at promoters that bind the HMGB family member Hmo1, and is reduced but not eliminated in hmo1Δ yeast. Finally, we show a reduced dependence on PIC components for Mediator occupancy at promoters after heat shock, further supporting altered dynamics or stronger engagement with activators under these conditions.
© 2022 Sarkar et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




Similar articles
-
Mediator, TATA-binding protein, and RNA polymerase II contribute to low histone occupancy at active gene promoters in yeast.J Biol Chem. 2014 May 23;289(21):14981-95. doi: 10.1074/jbc.M113.529354. Epub 2014 Apr 11. J Biol Chem. 2014. PMID: 24727477 Free PMC article.
-
Role of Mediator in regulating Pol II elongation and nucleosome displacement in Saccharomyces cerevisiae.Genetics. 2012 May;191(1):95-106. doi: 10.1534/genetics.111.135806. Epub 2012 Feb 29. Genetics. 2012. PMID: 22377631 Free PMC article.
-
Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and mediator tail subunits Med15 and Med16.J Biol Chem. 2013 Apr 26;288(17):12197-213. doi: 10.1074/jbc.M112.449553. Epub 2013 Feb 27. J Biol Chem. 2013. PMID: 23447536 Free PMC article.
-
Origins and activity of the Mediator complex.Semin Cell Dev Biol. 2011 Sep;22(7):729-34. doi: 10.1016/j.semcdb.2011.07.021. Epub 2011 Jul 28. Semin Cell Dev Biol. 2011. PMID: 21821140 Free PMC article. Review.
-
Toward understanding of the mechanisms of Mediator function in vivo: Focus on the preinitiation complex assembly.Transcription. 2017;8(5):328-342. doi: 10.1080/21541264.2017.1329000. Epub 2017 Aug 25. Transcription. 2017. PMID: 28841352 Free PMC article. Review.
Cited by
-
Moesin contributes to heat shock gene response through direct binding to the Med15 subunit of the Mediator complex in the nucleus.Open Biol. 2024 Oct;14(10):240110. doi: 10.1098/rsob.240110. Epub 2024 Oct 2. Open Biol. 2024. PMID: 39353569 Free PMC article.
-
Reliance of Host-Encoded Regulators of Retromobility on Ty1 Promoter Activity or Architecture.Front Mol Biosci. 2022 Jul 1;9:896215. doi: 10.3389/fmolb.2022.896215. eCollection 2022. Front Mol Biosci. 2022. PMID: 35847981 Free PMC article.
-
Function and dynamics of the Mediator complex: novel insights and new frontiers.Transcription. 2022 Feb-Jun;13(1-3):39-52. doi: 10.1080/21541264.2022.2085502. Epub 2022 Jun 16. Transcription. 2022. PMID: 35708525 Free PMC article. Review.
-
Growth-regulated co-occupancy of Mediator and Lsm3 at intronic ribosomal protein genes.Nucleic Acids Res. 2024 Jun 24;52(11):6220-6233. doi: 10.1093/nar/gkae266. Nucleic Acids Res. 2024. PMID: 38613396 Free PMC article.
-
Advancing the Indian cattle pangenome: characterizing non-reference sequences in Bos indicus.J Anim Sci Biotechnol. 2025 Feb 7;16(1):21. doi: 10.1186/s40104-024-01133-1. J Anim Sci Biotechnol. 2025. PMID: 39915889 Free PMC article.
References
-
- Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ, Berk AJ, Bjorklund S, Blackwell TK, Borggrefe T, Carey M, Carlson M, et al. 2004. A unified nomenclature for protein subunits of Mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell 14: 553–557. 10.1016/j.molcel.2004.05.011 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
