Serine palmitoyltransferase assembles at ER-mitochondria contact sites
- PMID: 34785538
- PMCID: PMC8605320
- DOI: 10.26508/lsa.202101278
Serine palmitoyltransferase assembles at ER-mitochondria contact sites
Abstract
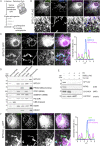
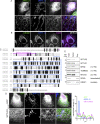
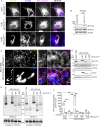
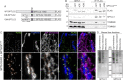
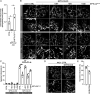
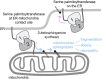
The accumulation of sphingolipid species in the cell contributes to the development of obesity and neurological disease. However, the subcellular localization of sphingolipid-synthesizing enzymes is unclear, limiting the understanding of where and how these lipids accumulate inside the cell and why they are toxic. Here, we show that SPTLC2, a subunit of the serine palmitoyltransferase (SPT) complex, catalyzing the first step in de novo sphingolipid synthesis, localizes dually to the ER and the outer mitochondrial membrane. We demonstrate that mitochondrial SPTLC2 interacts and forms a complex in trans with the ER-localized SPT subunit SPTLC1. Loss of SPTLC2 prevents the synthesis of mitochondrial sphingolipids and protects from palmitate-induced mitochondrial toxicity, a process dependent on mitochondrial ceramides. Our results reveal the in trans assembly of an enzymatic complex at an organellar membrane contact site, providing novel insight into the localization of sphingolipid synthesis and the composition and function of ER-mitochondria contact sites.
© 2021 Aaltonen et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
