Why are some coronavirus variants more infectious?
- PMID: 34785628
- PMCID: PMC8594289
- DOI: 10.1007/s12038-021-00221-y
Why are some coronavirus variants more infectious?
Abstract
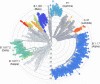
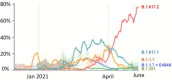
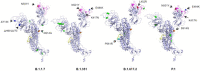
Since the start of the pandemic, SARS-CoV-2 has infected almost 200 million human hosts and is set to encounter and gain entry in many more in the coming months. As the coronavirus flourish, the evolutionary pressure selects those variants that can complete the infection cycle faster and reproduce in large numbers compared to others. This increase in infectivity and transmissibility coupled with the immune response from high viral load may cause moderate to severe disease. Whether this leads to enhanced virulence in the prevalent Alpha and Delta variants is still not clear. This review describes the different types of SARS-CoV-2 variants that are now prevalent, their emergence, the mutations responsible for their growth advantages, and how they affect vaccine efficacy and increase chances of reinfection. Finally, we have also summarized the efforts made to recognize and predict the mutations, which can cause immune escape and track their emergence through impactful genomic surveillance.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
