Modulation of AggR levels reveals features of virulence regulation in enteroaggregative E. coli
- PMID: 34785760
- PMCID: PMC8595720
- DOI: 10.1038/s42003-021-02820-9
Modulation of AggR levels reveals features of virulence regulation in enteroaggregative E. coli
Abstract
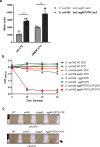
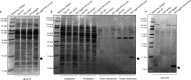
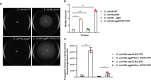
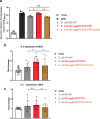
Enteroaggregative Escherichia coli (EAEC) strains are one of the diarrheagenic pathotypes. EAEC strains harbor a virulence plasmid (pAA2) that encodes, among other virulence determinants, the aggR gene. The expression of the AggR protein leads to the expression of several virulence determinants in both plasmids and chromosomes. In this work, we describe a novel mechanism that influences AggR expression. Because of the absence of a Rho-independent terminator in the 3'UTR, aggR transcripts extend far beyond the aggR ORF. These transcripts are prone to PNPase-mediated degradation. Structural alterations in the 3'UTR result in increased aggR transcript stability, leading to increased AggR levels. We therefore investigated the effect of increased AggR levels on EAEC virulence. Upon finding the previously described AggR-dependent virulence factors, we detected novel AggR-regulated genes that may play relevant roles in EAEC virulence. Mutants exhibiting high AggR levels because of structural alterations in the aggR 3'UTR show increased mobility and increased pAA2 conjugation frequency. Furthermore, among the genes exhibiting increased fold change values, we could identify those of metabolic pathways that promote increased degradation of arginine, fatty acids and gamma-aminobutyric acid (GABA), respectively. In this paper, we discuss how the AggR-dependent increase in specific metabolic pathways activity may contribute to EAEC virulence.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004;2:123–140. - PubMed
-
- Bhan MK, et al. Enteroaggregative Escherichia coli Associated with Persistent Diarrhea in a Cohort of Rural Children in India. J. Infect. Dis. 1989;159:1061–1064. - PubMed
-
- Fang GD, Lima AAM, Martins CV, Nataro JP, Guerrant RL. Etiology and epidemiology of persistent diarrhea in northeastern Brazil: a hospital-based, prospective, case-control study. J. Pediatr. Gastroenterol. Nutr. 1995;21:137–144. - PubMed

