Overexpressed GNA13 induces temozolomide sensitization via down-regulating MGMT and p-RELA in glioma
- PMID: 34786068
- PMCID: PMC8581860
Overexpressed GNA13 induces temozolomide sensitization via down-regulating MGMT and p-RELA in glioma
Abstract
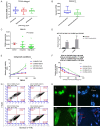
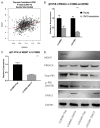
Temozolomide (TMZ), one of the few effective drugs used during adjuvant therapy, could effectively prolong the overall survival (OS) of glioma patients. In our previous study, the mRNA level of G Protein Subunit Alpha 13 (GNA13) was found to be inversely correlated with OS and was therefore identified as a potential biomarker for the prognosis of glioma. Henceforth, this study aims to identify the molecular mechanism of GNA13 in enhancing TMZ sensitization through bioinformatic analyses of GSE80729 and GSE43452 and other experiments. In glioma, overexpression of GNA13 downregulated PRKACA, which is a subunit of PKA, hence reducing phosphorylated RELA and MGMT. Since p-RELA and MGMT were proven to be closely associated with TMZ resistance, we therefore investigated whether thetwo signaling pathways, "GNA13/PRKACA/p-RELA", and "GNA13/PRKACA/MGMT", were involved in the molecular mechanism of GNA13 in TMZ sensitization. Our conclusion was that, GNA13 overexpression in glioma cells were more sensitive in TMZ treatment.
Keywords: GNA13; MGMT; PRKACA; RELA; glioblastoma; glioma; temozolomide (TMZ).
AJTR Copyright © 2021.
Conflict of interest statement
None.
Figures



References
-
- O’Hayre M, Inoue A, Kufareva I, Wang Z, Mikelis CM, Drummond RA, Avino S, Finkel K, Kalim KW, DiPasquale G, Guo F, Aoki J, Zheng Y, Lionakis MS, Molinolo AA, Gutkind JS. Inactivating mutations in GNA13 and RHOA in Burkitt’s lymphoma and diffuse large B-cell lymphoma: a tumor suppressor function for the Galpha13/RhoA axis in B cells. Oncogene. 2016;35:3771–80. - PMC - PubMed
-
- Rasheed SAK, Leong HS, Lakshmanan M, Raju A, Dadlani D, Chong FT, Shannon NB, Rajarethinam R, Skanthakumar T, Tan EY, Hwang JSG, Lim KH, Tan DS, Ceppi P, Wang M, Tergaonkar V, Casey PJ, Iyer NG. GNA13 expression promotes drug resistance and tumor-initiating phenotypes in squamous cell cancers. Oncogene. 2018;37:1340–53. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
