Identification of key genes and pathways in mild and severe nonalcoholic fatty liver disease by integrative analysis
- PMID: 34786546
- PMCID: PMC8579024
- DOI: 10.1016/j.cdtm.2021.08.002
Identification of key genes and pathways in mild and severe nonalcoholic fatty liver disease by integrative analysis
Abstract
Background: The global prevalence of nonalcoholic fatty liver disease (NAFLD) is increasing. The pathogenesis of NAFLD is multifaceted, and the underlying mechanisms are elusive. We conducted data mining analysis to gain a better insight into the disease and to identify the hub genes associated with the progression of NAFLD.
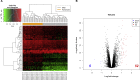
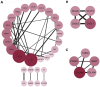
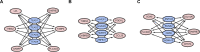
Methods: The dataset GSE49541, containing the profile of 40 samples representing mild stages of NAFLD and 32 samples representing advanced stages of NAFLD, was acquired from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) were identified using the R programming language. The Database for Annotation, Visualization and Integrated Discovery (DAVID) online tool and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database were used to perform the enrichment analysis and construct protein-protein interaction (PPI) networks, respectively. Subsequently, transcription factor networks and key modules were identified. The hub genes were validated in a mice model of high fat diet (HFD)-induced NAFLD and in cultured HepG2 cells by real-time quantitative PCR.
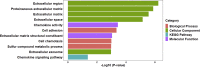
Results: Based on the GSE49541 dataset, 57 DEGs were selected and enriched in chemokine activity and cellular component, including the extracellular region. Twelve transcription factors associated with DEGs were indicated from PPI analysis. Upregulated expression of five hub genes (SOX9, CCL20, CXCL1, CD24, and CHST4), which were identified from the dataset, was also observed in the livers of HFD-induced NAFLD mice and in HepG2 cells exposed to palmitic acid or advanced glycation end products.
Conclusion: The hub genes SOX9, CCL20, CXCL1, CD24, and CHST4 are involved in the aggravation of NAFLD. Our results offer new insights into the underlying mechanism of NAFLD progression.
Keywords: Computational biology; Fatty liver; Nonalcoholic fatty liver disease.
© 2021 Chinese Medical Association. Publishing services by Elsevier B.V. on behalf of KeAi Communications Co. Ltd.
Conflict of interest statement
None.
Figures
Similar articles
-
Identification of hub genes and key pathways of dietary advanced glycation end products‑induced non‑alcoholic fatty liver disease by bioinformatics analysis and animal experiments.Mol Med Rep. 2020 Feb;21(2):685-694. doi: 10.3892/mmr.2019.10872. Epub 2019 Dec 9. Mol Med Rep. 2020. PMID: 31974594 Free PMC article.
-
Bioinformatics analysis reveals novel core genes associated with nonalcoholic fatty liver disease and nonalcoholic steatohepatitis.Gene. 2020 Jun 5;742:144549. doi: 10.1016/j.gene.2020.144549. Epub 2020 Mar 14. Gene. 2020. PMID: 32184169
-
Identification of genes and key pathways underlying the pathophysiological association between nonalcoholic fatty liver disease and atrial fibrillation.BMC Med Genomics. 2022 Jul 5;15(1):150. doi: 10.1186/s12920-022-01300-1. BMC Med Genomics. 2022. PMID: 35790963 Free PMC article.
-
WGCNA-Based Identification of Hub Genes and Key Pathways Involved in Nonalcoholic Fatty Liver Disease.Biomed Res Int. 2021 Dec 13;2021:5633211. doi: 10.1155/2021/5633211. eCollection 2021. Biomed Res Int. 2021. PMID: 34938809 Free PMC article.
-
Candidate genes investigation for severe nonalcoholic fatty liver disease based on bioinformatics analysis.Medicine (Baltimore). 2017 Aug;96(32):e7743. doi: 10.1097/MD.0000000000007743. Medicine (Baltimore). 2017. PMID: 28796060 Free PMC article.
Cited by
-
Transcriptome Sequencing Reveals Autophagy Networks in Rat Livers during the Development of NAFLD and Identifies Autophagy Hub Genes.Int J Mol Sci. 2023 Mar 29;24(7):6437. doi: 10.3390/ijms24076437. Int J Mol Sci. 2023. PMID: 37047411 Free PMC article.
-
Smad4 deficiency in hepatocytes attenuates NAFLD progression via inhibition of lipogenesis and macrophage polarization.Cell Death Dis. 2025 Jan 31;16(1):58. doi: 10.1038/s41419-025-07376-8. Cell Death Dis. 2025. PMID: 39890803 Free PMC article.
-
A regulatory element associated to NAFLD in the promoter of DIO1 controls LDL-C, HDL-C and triglycerides in hepatic cells.Lipids Health Dis. 2024 Feb 16;23(1):48. doi: 10.1186/s12944-024-02029-9. Lipids Health Dis. 2024. PMID: 38365720 Free PMC article.
-
Saturated Fat-Mediated Upregulation of IL-32 and CCL20 in Hepatocytes Contributes to Higher Expression of These Fibrosis-Driving Molecules in MASLD.Int J Mol Sci. 2023 Aug 25;24(17):13222. doi: 10.3390/ijms241713222. Int J Mol Sci. 2023. PMID: 37686029 Free PMC article.
-
Single-cell metabolic profiling reveals subgroups of primary human hepatocytes with heterogeneous responses to drug challenge.Genome Biol. 2023 Oct 17;24(1):234. doi: 10.1186/s13059-023-03075-9. Genome Biol. 2023. PMID: 37848949 Free PMC article.
References
-
- European Association for the Study of the Liver, European Association for the Study of Diabetes, European Association for the Study of Obesity. EASL-EASD-EASO clinical practice guidelines for the management of non-alcoholic fatty liver disease. J Hepatol. 2016;64:1388–1402. doi: 10.1007/s00125-016-3902-y. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

