Antimicrobial Resistance in Enterococcus Spp. Isolated from a Beef Processing Plant and Retail Ground Beef
- PMID: 34787441
- PMCID: PMC8597637
- DOI: 10.1128/Spectrum.01980-21
Antimicrobial Resistance in Enterococcus Spp. Isolated from a Beef Processing Plant and Retail Ground Beef
Abstract
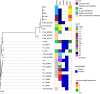
Antimicrobial use in food-producing animals has come under increasing scrutiny due to its potential association with antimicrobial resistance (AMR). Monitoring of AMR in indicator microorganisms such as Enterococcus spp. in meat production facilities and retail meat products can provide important information on the dynamics and prevalence of AMR in these environments. In this study, swabs or samples were obtained from various locations in a commercial beef packing operation (n = 600) and from retail ground beef (n = 60) over a 19-month period. All samples/swabs were enriched for Enterococcus spp., and suspected enterococci isolates were identified using species-specific PCR primers. Enterococcus faecalis was the most frequently isolated species, followed by Enterococcus hirae, which was found mostly on post-hide removal carcasses and in ground beef. Enterococcus faecium (n = 9) and E. faecalis (n = 120) isolates were further characterized for AMR. Twenty-one unique AMR profiles were identified, with 90% of isolates resistant to at least two antimicrobials and two that were resistant to nine antimicrobials. Tetracycline resistance was observed most often in E. faecalis (28.8%) and was likely mediated by tet(M). Genomic analysis of selected E. faecalis and E. faecium isolates revealed that many of the isolates in this study clustered with other publicly available genomes from ground beef, suggesting that these strains are well adapted to the beef processing environment. IMPORTANCE Antimicrobial resistance (AMR) is a serious challenge facing the agricultural industry. Understanding the flow of antimicrobial-resistant bacteria through the beef fabrication process and into ground beef is an important step in identifying intervention points for reducing AMR. In this study, we used enterococci as indicator bacteria for monitoring AMR in a commercial beef packaging facility and in retail ground beef over a 19-month period. Although washing of carcasses post-hide removal reduced the isolation frequency of Enterococcus spp., a number of antimicrobial-resistant Enterococcus faecalis isolates were recovered from ground beef produced in the packaging plant. Genome analysis showed that several E. faecalis isolates were genetically similar to publicly available isolates recovered from retail ground beef in the United States.
Keywords: Enterococcus; abattoir; antimicrobial resistance; beef; multidrug resistance.
Figures


References
-
- Boehm AB, Sassoubre LM. 2014. Enterococci as indicators of environmental fecal contamination. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed) Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. - PubMed
-
- Farnleitner AH, Ryzinska-Paier G, Reischer GH, Burtscher MM, Knetsch S, Kirschner AK, Dirnbock T, Kuschnig G, Mach RL, Sommer R. 2010. Escherichia coli and enterococci are sensitive and reliable indicators for human, livestock and wildlife faecal pollution in alpine mountainous water resources. J Appl Microbiol 109:1599–1608. doi: 10.1111/j.1365-2672.2010.04788.x. - DOI - PMC - PubMed
-
- Cernicchiaro N, Oliveira ARS, Hoehn A, Cull CA, Noll LW, Shridhar PB, Nagaraja TG, Ives SE, Renter DG, Sanderson MW. 2019. Quantification of bacteria indicative of fecal and environmental contamination from hides to carcasses. Foodborne Pathog Dis 16:844–855. doi: 10.1089/fpd.2019.2656. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

