Prolonged SARS-CoV-2 Positivity in Immunocompetent Patients: Virus Isolation, Genomic Integrity, and Transmission Risk
- PMID: 34787498
- PMCID: PMC8597635
- DOI: 10.1128/Spectrum.00855-21
Prolonged SARS-CoV-2 Positivity in Immunocompetent Patients: Virus Isolation, Genomic Integrity, and Transmission Risk
Abstract
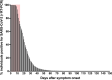
Current guidelines for patient isolation in COVID-19 cases recommend a symptom-based approach, averting the use of control real-time reverse transcription PCR (rRT-PCR) testing. However, we hypothesized that patients with persistently positive results by RT-PCR for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) could be potentially infectious for a prolonged time, even if immunocompetent and asymptomatic, which would demand a longer social isolation period than presently recommended. To test this hypothesis, 72 samples from 51 mildly symptomatic immunocompetent patients with long-lasting positive rRT-PCR results for SARS-CoV-2 were tested for their infectiousness in cell culture. The serological response of samples from those patients and virus genomic integrity were also analyzed. Infectious viruses were successfully isolated from 34.38% (22/64) of nasopharynx samples obtained 14 days or longer after symptom onset. Indeed, we observed successful virus isolation up to 128 days. Complete SARS-COV-2 genome integrity was demonstrated, suggesting the presence of replication-competent viruses. No correlation was found between the isolation of infectious viruses and rRT-PCR cycle threshold values or the humoral immune response. These findings call attention to the need to review current isolation guidelines, particularly in scenarios involving high-risk individuals. IMPORTANCE In this study, we evaluated mildly symptomatic immunocompetent patients with long-lasting positive rRT-PCR results for SARS-CoV-2. Infectious viruses were successfully isolated in cell cultures from nasopharynx samples obtained 14 days or longer after symptom onset. Indeed, we observed successful virus isolation for up to 128 days. Moreover, SARS-CoV-2 genome integrity was demonstrated by sequencing, suggesting the presence of replication-competent viruses. These data point out the risk of continuous SARS-CoV-2 transmission from patients with prolonged detection of SARS-CoV-2 in the upper respiratory tract, which has important implications for current precaution guidelines, particularly in settings where vulnerable individuals may be exposed (e.g., nursing homes and hospitals).
Keywords: COVID-19; COVID-19 nucleic acid testing; SARS-CoV-2; immunocompetent; persistence; virus shedding.
Conflict of interest statement
We declare no conflicts of interest.
Figures


Similar articles
-
Shedding of Viable Virus in Asymptomatic SARS-CoV-2 Carriers.mSphere. 2021 May 19;6(3):e00019-21. doi: 10.1128/mSphere.00019-21. mSphere. 2021. PMID: 34011679 Free PMC article.
-
Persistent SARS-CoV-2 RNA Shedding Without Evidence of Infectiousness: A Cohort Study of Individuals With COVID-19.J Infect Dis. 2021 Oct 28;224(8):1362-1371. doi: 10.1093/infdis/jiab107. J Infect Dis. 2021. PMID: 33649773 Free PMC article.
-
Inference of Active Viral Replication in Cases with Sustained Positive Reverse Transcription-PCR Results for SARS-CoV-2.J Clin Microbiol. 2021 Jan 21;59(2):e02277-20. doi: 10.1128/JCM.02277-20. Print 2021 Jan 21. J Clin Microbiol. 2021. PMID: 33239378 Free PMC article.
-
Viral cultures, cycle threshold values and viral load estimation for assessing SARS-CoV-2 infectiousness in haematopoietic stem cell and solid organ transplant patients: a systematic review.J Hosp Infect. 2023 Feb;132:62-72. doi: 10.1016/j.jhin.2022.11.018. Epub 2022 Dec 5. J Hosp Infect. 2023. PMID: 36473552 Free PMC article.
-
Viral dynamics in the Upper Respiratory Tract (URT) of SARS-CoV-2.Infez Med. 2020 Dec 1;28(4):486-499. Infez Med. 2020. PMID: 33257622
Cited by
-
Which criteria should we use to end isolation in hemodialysis patients with COVID-19?Clin Kidney J. 2022 Apr 30;15(8):1450-1454. doi: 10.1093/ckj/sfac115. eCollection 2022 Aug. Clin Kidney J. 2022. PMID: 36824062 Free PMC article.
-
Post-market surveillance of six COVID-19 point-of-care tests using pre-Omicron and Omicron SARS-CoV-2 variants.Microbiol Spectr. 2024 Jul 2;12(7):e0016324. doi: 10.1128/spectrum.00163-24. Epub 2024 May 17. Microbiol Spectr. 2024. PMID: 38757955 Free PMC article.
-
Oligosymptomatic long-term carriers of SARS-CoV-2 display impaired innate resistance but increased high-affinity anti-spike antibodies.iScience. 2023 Jun 28;26(7):107219. doi: 10.1016/j.isci.2023.107219. eCollection 2023 Jul 21. iScience. 2023. PMID: 37529320 Free PMC article.
-
Immediate Postoperative COVID-19 Infection after Lung Transplantation: A Systematic Review and Case Series.J Clin Med. 2023 Nov 10;12(22):7028. doi: 10.3390/jcm12227028. J Clin Med. 2023. PMID: 38002643 Free PMC article. Review.
-
Cycle Threshold Values of SARS-CoV-2 RT-PCR during Outbreaks in Nursing Homes: A Retrospective Cohort Study.Epidemiologia (Basel). 2024 Oct 16;5(4):658-668. doi: 10.3390/epidemiologia5040046. Epidemiologia (Basel). 2024. PMID: 39449389 Free PMC article.
References
-
- World Health Organization. 2021. WHO coronavirus (COVID-19) dashboard. World Health Organization, Geneva, Switzerland. https://covid19.who.int/.
-
- Coronavirus Brasil. 2021. Painel coronavirus. https://covid.saude.gov.br.
-
- Vacinacaocovid-19.saude.rj. 2021. Campanha de Vacinação Covid-19. Secretaria de Saúde do Estado do Rio de Janeiro. https://vacinacaocovid19.saude.rj.gov.br/vacinometro.
-
- To KK-W, Tsang OT-Y, Leung W-S, Tam AR, Wu T-C, Lung DC, Yip CC-Y, Cai J-P, Chan JM-C, Chik TS-H, Lau DP-L, Choi CY-C, Chen L-L, Chan W-M, Chan K-H, Ip JD, Ng AC-K, Poon RW-S, Luo C-T, Cheng VC-C, Chan JF-W, Hung IF-N, Chen Z, Chen H, Yuen K-Y. 2020. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 20:565–574. doi:10.1016/S1473-3099(20)30196-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

