Whole set of constitutive promoters for RpoN sigma factor and the regulatory role of its enhancer protein NtrC in Escherichia coli K-12
- PMID: 34787538
- PMCID: PMC8743547
- DOI: 10.1099/mgen.0.000653
Whole set of constitutive promoters for RpoN sigma factor and the regulatory role of its enhancer protein NtrC in Escherichia coli K-12
Abstract
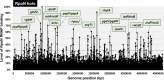
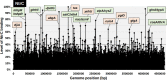
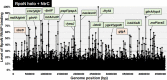
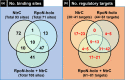
The promoter selectivity of Escherichia coli RNA polymerase (RNAP) is determined by its promoter-recognition sigma subunit. The model prokaryote E. coli K-12 contains seven species of the sigma subunit, each recognizing a specific set of promoters. Using genomic SELEX (gSELEX) screening in vitro, we identified the whole set of 'constitutive' promoters recognized by the reconstituted RNAP holoenzyme alone, containing RpoD (σ70), RpoS (σ38), RpoH (σ32), RpoF (σ28) or RpoE (σ24), in the absence of other supporting regulatory factors. In contrast, RpoN sigma (σ54), involved in expression of nitrogen-related genes and also other cellular functions, requires an enhancer (or activator) protein, such as NtrC, for transcription initiation. In this study, a series of gSELEX screenings were performed to search for promoters recognized by the RpoN RNAP holoenzyme in the presence and absence of the major nitrogen response enhancer NtrC, the best-characterized enhancer. Based on the RpoN holoenzyme-binding sites, a total of 44 to 61 putative promoters were identified, which were recognized by the RpoN holoenzyme alone. In the presence of the enhancer NtrC, the recognition target increased to 61-81 promoters. Consensus sequences of promoters recognized by RpoN holoenzyme in the absence and presence of NtrC were determined. The promoter activity of a set of NtrC-dependent and -independent RpoN promoters was verified in vivo under nitrogen starvation, in the presence and absence of RpoN and/or NtrC. The promoter activity of some RpoN-recognized promoters increased in the absence of RpoN or NtrC, supporting the concept that the promoter-bound NtrC-enhanced RpoN holoenzyme functions as a repressor against RpoD holoenzyme. Based on our findings, we propose a model in which the RpoN holoenzyme fulfils the dual role of repressor and transcriptase for the same set of genes. We also propose that the promoter recognized by RpoN holoenzyme in the absence of enhancers is the 'repressive' promoter. The presence of high-level RpoN sigma in growing E. coli K-12 in rich medium may be related to the repression role of a set of genes needed for the utilization of ammonia as a nitrogen source in poor media. The list of newly identified regulatory targets of RpoN provides insight into E. coli survival under nitrogen-depleted conditions in nature.
Keywords: Escherichia coli; NtrC; RNA polymerase; RpoN sigma factor; gSELEX; nitrogen metabolism.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


Similar articles
-
The whole set of the constitutive promoters recognized by four minor sigma subunits of Escherichia coli RNA polymerase.PLoS One. 2017 Jun 30;12(6):e0179181. doi: 10.1371/journal.pone.0179181. eCollection 2017. PLoS One. 2017. PMID: 28666008 Free PMC article.
-
The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli.PLoS One. 2014 Mar 6;9(3):e90447. doi: 10.1371/journal.pone.0090447. eCollection 2014. PLoS One. 2014. PMID: 24603758 Free PMC article.
-
Sigma factor N, liaison to an ntrC and rpoS dependent regulatory pathway controlling acid resistance and the LEE in enterohemorrhagic Escherichia coli.PLoS One. 2012;7(9):e46288. doi: 10.1371/journal.pone.0046288. Epub 2012 Sep 27. PLoS One. 2012. PMID: 23029465 Free PMC article.
-
The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences.FEMS Microbiol Lett. 2000 May 1;186(1):1-9. doi: 10.1111/j.1574-6968.2000.tb09074.x. FEMS Microbiol Lett. 2000. PMID: 10779705 Review.
-
Hierarchy of transcription factor network in Escherichia coli K-12: H-NS-mediated silencing and Anti-silencing by global regulators.FEMS Microbiol Rev. 2021 Nov 23;45(6):fuab032. doi: 10.1093/femsre/fuab032. FEMS Microbiol Rev. 2021. PMID: 34196371 Review.
Cited by
-
Genomic SELEX Screening of Regulatory Targets of Transcription Factors.Methods Mol Biol. 2024;2819:77-102. doi: 10.1007/978-1-0716-3930-6_5. Methods Mol Biol. 2024. PMID: 39028503
-
Cell-growth phase-dependent promoter replacement approach for improved poly(lactate-co-3-hydroxybutyrate) production in Escherichia coli.Microb Cell Fact. 2023 Jul 19;22(1):131. doi: 10.1186/s12934-023-02143-w. Microb Cell Fact. 2023. PMID: 37468909 Free PMC article.
-
A Role for the RNA Polymerase Gene Specificity Factor σ54 in the Uniform Colony Growth of Uropathogenic Escherichia coli.J Bacteriol. 2022 Apr 19;204(4):e0003122. doi: 10.1128/jb.00031-22. Epub 2022 Mar 31. J Bacteriol. 2022. PMID: 35357162 Free PMC article.
-
Characterization of the transcriptionally active form of dephosphorylated DctD complexed with dephospho-IIAGlc.mBio. 2024 May 8;15(5):e0033024. doi: 10.1128/mbio.00330-24. Epub 2024 Apr 2. mBio. 2024. PMID: 38564689 Free PMC article.
-
Expanded roles of lactate-sensing LldR in transcription regulation of the Escherichia coli K-12 genome: lactate utilisation and acid resistance.Microb Genom. 2023 May;9(5):mgen001015. doi: 10.1099/mgen.0.001015. Microb Genom. 2023. PMID: 37219924 Free PMC article.
References
-
- Hirschman J, Wong PK, Sei K, Keener J, Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci USA. 1985;82:7525–7529. doi: 10.1073/pnas.82.22.7525. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

