MicroRNAs affect GPCR and Ion channel genes needed for influenza replication
- PMID: 34787540
- PMCID: PMC8742985
- DOI: 10.1099/jgv.0.001691
MicroRNAs affect GPCR and Ion channel genes needed for influenza replication
Abstract
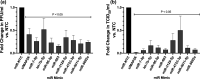
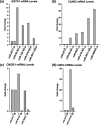
Influenza virus causes seasonal epidemics and sporadic pandemics resulting in morbidity, mortality, and economic losses worldwide. Understanding how to regulate influenza virus replication is important for developing vaccine and therapeutic strategies. Identifying microRNAs (miRs) that affect host genes used by influenza virus for replication can support an antiviral strategy. In this study, G-protein coupled receptor (GPCR) and ion channel (IC) host genes in human alveolar epithelial (A549) cells used by influenza virus for replication (Orr-Burks et al., 2021) were examined as miR target genes following A/CA/04/09- or B/Yamagata/16/1988 replication. Thirty-three miRs were predicted to target GPCR or IC genes and their miR mimics were evaluated for their ability to decrease influenza virus replication. Paired miR inhibitors were used as an ancillary measure to confirm or not the antiviral effects of a miR mimic. Fifteen miRs lowered influenza virus replication and four miRs were found to reduce replication irrespective of virus strain and type differences. These findings provide evidence for novel miR disease intervention strategies for influenza viruses.
Keywords: GPCR; influenza; ion channel; microRNA.
Figures
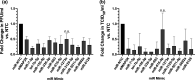
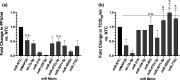

Similar articles
-
G-Protein-Coupled Receptor and Ion Channel Genes Used by Influenza Virus for Replication.J Virol. 2021 Apr 12;95(9):e02410-20. doi: 10.1128/JVI.02410-20. Print 2021 Apr 12. J Virol. 2021. PMID: 33536179 Free PMC article.
-
Identification of Host Kinase Genes Required for Influenza Virus Replication and the Regulatory Role of MicroRNAs.PLoS One. 2013 Jun 21;8(6):e66796. doi: 10.1371/journal.pone.0066796. Print 2013. PLoS One. 2013. PMID: 23805279 Free PMC article.
-
Enhancing the yield of seasonal influenza viruses through manipulation of microRNAs in Madin-Darby canine kidney cells.Exp Biol Med (Maywood). 2022 Aug;247(15):1335-1349. doi: 10.1177/15353702221098340. Epub 2022 Jun 6. Exp Biol Med (Maywood). 2022. PMID: 35666095 Free PMC article.
-
Roles of lncRNAs in influenza virus infection.Emerg Microbes Infect. 2020 Dec;9(1):1407-1414. doi: 10.1080/22221751.2020.1778429. Emerg Microbes Infect. 2020. PMID: 32543285 Free PMC article. Review.
-
Regulation of influenza virus infection by long non-coding RNAs.Virus Res. 2016 Jan 2;212:78-84. doi: 10.1016/j.virusres.2015.08.008. Epub 2015 Aug 29. Virus Res. 2016. PMID: 26321158 Review.
Cited by
-
Specific GPCRs Elicit Unique Extracellular Vesicle MiRNA Array Signatures: An Exploratory Study.bioRxiv [Preprint]. 2025 Jun 20:2025.06.16.659918. doi: 10.1101/2025.06.16.659918. bioRxiv. 2025. PMID: 40667070 Free PMC article. Preprint.
-
Exosomal mir-126-3p derived from endothelial cells induces ion channel dysfunction by targeting RGS3 signaling in cardiomyocytes: a novel mechanism in Takotsubo cardiomyopathy.Stem Cell Res Ther. 2025 Feb 4;16(1):36. doi: 10.1186/s13287-025-04157-0. Stem Cell Res Ther. 2025. PMID: 39901299 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

