Accuracies of genomic predictions for disease resistance of striped catfish to Edwardsiella ictaluri using artificial intelligence algorithms
- PMID: 34788431
- PMCID: PMC8727988
- DOI: 10.1093/g3journal/jkab361
Accuracies of genomic predictions for disease resistance of striped catfish to Edwardsiella ictaluri using artificial intelligence algorithms
Abstract
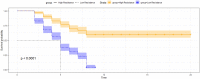
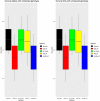
Assessments of genomic prediction accuracies using artificial intelligent (AI) algorithms (i.e., machine and deep learning methods) are currently not available or very limited in aquaculture species. The principal aim of this study was to examine the predictive performance of these new methods for disease resistance to Edwardsiella ictaluri in a population of striped catfish Pangasianodon hypophthalmus and to make comparisons with four common methods, i.e., pedigree-based best linear unbiased prediction (PBLUP), genomic-based best linear unbiased prediction (GBLUP), single-step GBLUP (ssGBLUP) and a nonlinear Bayesian approach (notably BayesR). Our analyses using machine learning (i.e., ML-KAML) and deep learning (i.e., DL-MLP and DL-CNN) together with the four common methods (PBLUP, GBLUP, ssGBLUP, and BayesR) were conducted for two main disease resistance traits (i.e., survival status coded as 0 and 1 and survival time, i.e., days that the animals were still alive after the challenge test) in a pedigree consisting of 560 individual animals (490 offspring and 70 parents) genotyped for 14,154 single nucleotide polymorphism (SNPs). The results using 6,470 SNPs after quality control showed that machine learning methods outperformed PBLUP, GBLUP, and ssGBLUP, with the increases in the prediction accuracies for both traits by 9.1-15.4%. However, the prediction accuracies obtained from machine learning methods were comparable to those estimated using BayesR. Imputation of missing genotypes using AlphaFamImpute increased the prediction accuracies by 5.3-19.2% in all the methods and data used. On the other hand, there were insignificant decreases (0.3-5.6%) in the prediction accuracies for both survival status and survival time when multivariate models were used in comparison to univariate analyses. Interestingly, the genomic prediction accuracies based on only highly significant SNPs (P < 0.00001, 318-400 SNPs for survival status and 1,362-1,589 SNPs for survival time) were somewhat lower (0.3-15.6%) than those obtained from the whole set of 6,470 SNPs. In most of our analyses, the accuracies of genomic prediction were somewhat higher for survival time than survival status (0/1 data). It is concluded that although there are prospects for the application of genomic selection to increase disease resistance to E. ictaluri in striped catfish breeding programs, further evaluation of these methods should be made in independent families/populations when more data are accumulated in future generations to avoid possible biases in the genetic parameters estimates and prediction accuracies for the disease-resistant traits studied in this population of striped catfish P. hypophthalmus.
Keywords: Edwardsiella ictaluri; Pangasianodon hypophthalmus; BNP disease; BayesR; genomic prediction; machine learning and deep learning; striped catfish.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures

References
-
- Abdollahi-Arpanahi R, Morota G, Peñagaricano F.. 2017. Predicting bull fertility using genomic data and biological information. J Dairy Sci. 100:9656–9666. - PubMed
-
- Aguilar I, Misztal I, Johnson D, Legarra A, Tsuruta S, et al.2010. Hot topic: a unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J Dairy Sci. 93:743–752. - PubMed
-
- Bargelloni L, Tassiello O, Babbucci M, Ferraresso S, Franch R, et al.2021. Data imputation and machine learning improve association analysis and genomic prediction for resistance to fish photobacteriosis in the gilthead sea bream. Aquacul Rep. 20:100661.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
