GADGETS: a genetic algorithm for detecting epistasis using nuclear families
- PMID: 34788792
- PMCID: PMC10060691
- DOI: 10.1093/bioinformatics/btab766
GADGETS: a genetic algorithm for detecting epistasis using nuclear families
Erratum in
-
Erratum to: GADGETS: a genetic algorithm for detecting epistasis using nuclear families.Bioinformatics. 2022 Jan 12;38(3):881. doi: 10.1093/bioinformatics/btab833. Bioinformatics. 2022. PMID: 34950948 Free PMC article. No abstract available.
Abstract
Motivation: Epistasis may play an etiologic role in complex diseases, but research has been hindered because identification of interactions among sets of single nucleotide polymorphisms (SNPs) requires exploration of immense search spaces. Current approaches using nuclear families accommodate at most several hundred candidate SNPs.
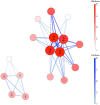
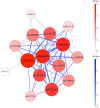
Results: GADGETS detects epistatic SNP-sets by applying a genetic algorithm to case-parent or case-sibling data. To allow for multiple epistatic sets, island subpopulations of SNP-sets evolve separately under selection for evident joint relevance to disease risk. The software evaluates the identified SNP-sets via permutation testing and provides graphical visualization. GADGETS correctly identified epistatic SNP-sets in realistically simulated case-parent triads with 10 000 candidate SNPs, far more SNPs than competitors can handle, and it outperformed competitors in simulations with many fewer SNPs. Applying GADGETS to family-based oral-clefting data from dbGaP identified SNP-sets with possible epistatic effects on risk.
Availability and implementation: GADGETS is part of the epistasisGA package at https://github.com/mnodzenski/epistasisGA.
Supplementary information: Supplementary data are available at Bioinformatics online.
Published by Oxford University Press 2021. This work is written by a US Government employee and is in the public domain in the US.
Figures


References
-
- Andre J.H., Koza J.R. (1996). Parallel genetic programming: a scalable implementation using the transputer network architecture. In: Angeline P.J., Kinnear K.E. (eds.), Advances in Genetic Programming, Vol. 2, Chap. 16. MIT Press, Cambridge, MA.

