Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders
- PMID: 34788896
- PMCID: PMC9299702
- DOI: 10.1002/chem.202103520
Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders
Abstract
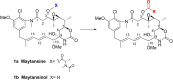
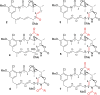
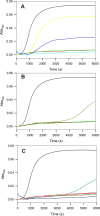
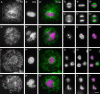
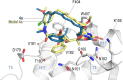
Maytansinol is a valuable precursor for the preparation of maytansine derivatives (known as maytansinoids). Inspired by the intriguing structure of the macrocycle and the success in targeted cancer therapy of the derivatives, we explored the maytansinol acylation reaction. As a result, we were able to obtain a series of derivatives with novel modifications of the maytansine scaffold. We characterized these molecules by docking studies, by a comprehensive biochemical evaluation, and by determination of their crystal structures in complex with tubulin. The results shed further light on the intriguing chemical behavior of maytansinoids and confirm the relevance of this peculiar scaffold in the scenario of tubulin binders.
Keywords: maytansine binding site; maytansinol; microtubules; tubulin; tubulin binders.
© 2021 The Authors. Chemistry - A European Journal published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
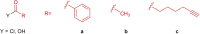



References
-
- Kupchan S. M., Komoda Y., Court W. A., Thomas G. J., Smith R. M., Karim A., Gilmore C. J., Haitiwanger R. C., Bryan R. F., J. Am. Chem. Soc. 1972, 1354. - PubMed
-
- Remillard S., Rebhun L. I., Howie G. A., Kupchan S. M., Science 1975, 189, 1002–1005. - PubMed
-
- Issell B. F., Crooke S. T., Cancer Treat. Rev. 1978, 5, 199–207. - PubMed
-
- Cassady J. M., Chan K. K., Floss H. G., Leistner E., Chem. Pharm. Bull. 2004, 52, 1–26. - PubMed

