Comprehensive characterization of copy number variation (CNV) called from array, long- and short-read data
- PMID: 34789167
- PMCID: PMC8596897
- DOI: 10.1186/s12864-021-08082-3
Comprehensive characterization of copy number variation (CNV) called from array, long- and short-read data
Abstract
Background: SNP arrays, short- and long-read genome sequencing are genome-wide high-throughput technologies that may be used to assay copy number variants (CNVs) in a personal genome. Each of these technologies comes with its own limitations and biases, many of which are well-known, but not all of them are thoroughly quantified.
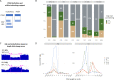
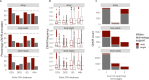
Results: We assembled an ensemble of public datasets of published CNV calls and raw data for the well-studied Genome in a Bottle individual NA12878. This assembly represents a variety of methods and pipelines used for CNV calling from array, short- and long-read technologies. We then performed cross-technology comparisons regarding their ability to call CNVs. Different from other studies, we refrained from using the golden standard. Instead, we attempted to validate the CNV calls by the raw data of each technology.
Conclusions: Our study confirms that long-read platforms enable recalling CNVs in genomic regions inaccessible to arrays or short reads. We also found that the reproducibility of a CNV by different pipelines within each technology is strongly linked to other CNV evidence measures. Importantly, the three technologies show distinct public database frequency profiles, which differ depending on what technology the database was built on.
Keywords: CNV; Genome in a Bottle; Long reads; Microarrays; Short reads.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, Kussmann J, Shashi V, Johnson K, Rehder C, Ballif BC, Shaffer LG, Eichler EE. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43(9):838–46. - PMC - PubMed
-
- Mace A, Tuke MA, Deelen P, Kristiansson K, Mattsson H, Noukas M, Sapkota Y, Schick U, Porcu E, Rueger S, McDaid AF, Porteous D, Winkler TW, Salvi E, Shrine N, Liu X, Ang WQ, Zhang W, Feitosa MF, Venturini C, van der Most PJ, Rosengren A, Wood AR, Beaumont RN, Jones SE, Ruth KS, Yaghootkar H, Tyrrell J, Havulinna AS, Boers H, Magi R, Kriebel J, Muller-Nurasyid M, Perola M, Nieminen M, Lokki ML, Kahonen M, Viikari JS, Geller F, Lahti J, Palotie A, Koponen P, Lundqvist A, Rissanen H, Bottinger EP, Afaq S, Wojczynski MK, Lenzini P, Nolte IM, Sparso T, Schupf N, Christensen K, Perls TT, Newman AB, Werge T, Snieder H, Spector TD, Chambers JC, Koskinen S, Melbye M, Raitakari OT, Lehtimaki T, Tobin MD, Wain LV, Sinisalo J, Peters A, Meitinger T, Martin NG, Wray NR, Montgomery GW, Medland SE, Swertz MA, Vartiainen E, Borodulin K, Mannisto S, Murray A, Bochud M, Jacquemont S, Rivadeneira F, Hansen TF, Oldehinkel AJ, Mangino M, Province MA, Deloukas P, Kooner JS, Freathy RM, Pennell C, Feenstra B, Strachan DP, Lettre G, Hirschhorn J, Cusi D, Heid IM, Hayward C, Mannik K, Beckmann JS, Loos RJF, Nyholt DR, Metspalu A, Eriksson JG, et al. Cnv-association meta-analysis in 191,161 european adults reveals new loci associated with anthropometric traits. Nat Commun. 2017;8(1):744. - PMC - PubMed
-
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–51. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

