Sexually dimorphic patterns in maternal circulating microRNAs in pregnancies complicated by fetal growth restriction
- PMID: 34789323
- PMCID: PMC8597318
- DOI: 10.1186/s13293-021-00405-z
Sexually dimorphic patterns in maternal circulating microRNAs in pregnancies complicated by fetal growth restriction
Abstract
Background: Current methods fail to accurately predict women at greatest risk of developing fetal growth restriction (FGR) or related adverse outcomes, including stillbirth. Sexual dimorphism in these adverse pregnancy outcomes is well documented as are sex-specific differences in gene and protein expression in the placenta. Circulating maternal serum microRNAs (miRNAs) offer potential as biomarkers that may also be informative of underlying pathology. We hypothesised that FGR would be associated with an altered miRNA profile and would differ depending on fetal sex.
Methods: miRNA expression profiles were assessed in maternal serum (> 36 weeks' gestation) from women delivering a severely FGR infant (defined as an individualised birthweight centile (IBC) < 3rd) and matched control participants (AGA; IBC = 20-80th), using miRNA arrays. qPCR was performed using specific miRNA primers in an expanded cohort of patients with IBC < 5th (n = 15 males, n = 16 females/group). Maternal serum human placental lactogen (hPL) was used as a proxy to determine if serum miRNAs were related to placental dysfunction. In silico analyses were performed to predict the potential functions of altered miRNAs.
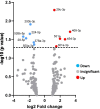
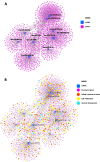
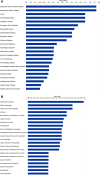
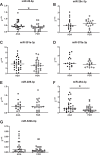
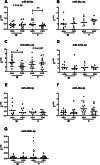
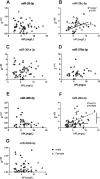
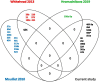
Results: Initial analyses revealed 11 miRNAs were altered in maternal serum from FGR pregnancies. In silico analyses revealed all 11 altered miRNAs were located in a network of genes that regulate placental function. Subsequent analysis demonstrated four miRNAs showed sexually dimorphic patterns. miR-28-5p was reduced in FGR pregnancies (p < 0.01) only when there was a female offspring and miR-301a-3p was only reduced in FGR pregnancies with a male fetus (p < 0.05). miR-454-3p was decreased in FGR pregnancies (p < 0.05) regardless of fetal sex but was only positively correlated to hPL when the fetus was female. Conversely, miR-29c-3p was correlated to maternal hPL only when the fetus was male. Target genes for sexually dimorphic miRNAs reveal potential functional roles in the placenta including angiogenesis, placental growth, nutrient transport and apoptosis.
Conclusions: These studies have identified sexually dimorphic patterns for miRNAs in maternal serum in FGR. These miRNAs may have potential as non-invasive biomarkers for FGR and associated placental dysfunction. Further studies to determine if these miRNAs have potential functional roles in the placenta may provide greater understanding of the pathogenesis of placental dysfunction and the differing susceptibility of male and female fetuses to adverse in utero conditions.
Keywords: Biomarker; FGR; Placenta; Placental dysfunction; Pregnancy; Serum; Sexual dimorphism; Stillbirth; miRNA.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no commercial or financial competing interests.
Figures

Similar articles
-
Identifying late-onset fetal growth restriction by measuring circulating placental RNA in the maternal blood at 28 weeks' gestation.Am J Obstet Gynecol. 2016 Apr;214(4):521.e1-521.e8. doi: 10.1016/j.ajog.2016.01.191. Epub 2016 Feb 12. Am J Obstet Gynecol. 2016. PMID: 26880734
-
Biochemical tests of placental function versus ultrasound assessment of fetal size for stillbirth and small-for-gestational-age infants.Cochrane Database Syst Rev. 2019 May 14;5(5):CD012245. doi: 10.1002/14651858.CD012245.pub2. Cochrane Database Syst Rev. 2019. PMID: 31087568 Free PMC article.
-
The effect of maternal position on placental blood flow and fetoplacental oxygenation in late gestation fetal growth restriction: a magnetic resonance imaging study.J Physiol. 2023 Dec;601(23):5391-5411. doi: 10.1113/JP284269. Epub 2023 Jul 19. J Physiol. 2023. PMID: 37467072
-
First trimester prediction models for small-for- gestational age and fetal growth restricted fetuses without the presence of preeclampsia.Mol Cell Probes. 2023 Dec;72:101941. doi: 10.1016/j.mcp.2023.101941. Epub 2023 Nov 16. Mol Cell Probes. 2023. PMID: 37951512
-
Mechanistic insights into the development of severe fetal growth restriction.Clin Sci (Lond). 2023 Apr 26;137(8):679-695. doi: 10.1042/CS20220284. Clin Sci (Lond). 2023. PMID: 37186255 Free PMC article. Review.
Cited by
-
Impact of placental pathology on the risk of bronchopulmonary dysplasia in preterm infants: The role of gestational age and sex.Eur J Pediatr. 2025 Feb 26;184(3):211. doi: 10.1007/s00431-025-06016-9. Eur J Pediatr. 2025. PMID: 40009187
-
The role of microRNAs in pregnancies complicated by maternal diabetes.Clin Sci (Lond). 2024 Sep 18;138(18):1179-1207. doi: 10.1042/CS20230681. Clin Sci (Lond). 2024. PMID: 39289953 Free PMC article. Review.
-
Modelling human placental villous development: designing cultures that reflect anatomy.Cell Mol Life Sci. 2022 Jun 26;79(7):384. doi: 10.1007/s00018-022-04407-x. Cell Mol Life Sci. 2022. PMID: 35753002 Free PMC article. Review.
-
Clinical value of prenatal screening markers in early pregnancy combined with perinatal characteristics to predict fetal growth restriction.Transl Pediatr. 2024 Jul 31;13(7):1071-1085. doi: 10.21037/tp-24-58. Epub 2024 Jul 29. Transl Pediatr. 2024. PMID: 39144423 Free PMC article.
-
Differential Expression of Maternal Plasma microRNAs and Their Respective Gene Targets Can Predict Early Fetal Growth Restriction.Life (Basel). 2025 Jan 24;15(2):167. doi: 10.3390/life15020167. Life (Basel). 2025. PMID: 40003576 Free PMC article. Review.
References
-
- Statistics OFN. Births in England and Wales: 2018. 2019.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

