Seasonal mixed layer depth shapes phytoplankton physiology, viral production, and accumulation in the North Atlantic
- PMID: 34789722
- PMCID: PMC8599477
- DOI: 10.1038/s41467-021-26836-1
Seasonal mixed layer depth shapes phytoplankton physiology, viral production, and accumulation in the North Atlantic
Abstract
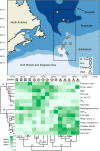
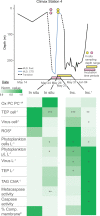
Seasonal shifts in phytoplankton accumulation and loss largely follow changes in mixed layer depth, but the impact of mixed layer depth on cell physiology remains unexplored. Here, we investigate the physiological state of phytoplankton populations associated with distinct bloom phases and mixing regimes in the North Atlantic. Stratification and deep mixing alter community physiology and viral production, effectively shaping accumulation rates. Communities in relatively deep, early-spring mixed layers are characterized by low levels of stress and high accumulation rates, while those in the recently shallowed mixed layers in late-spring have high levels of oxidative stress. Prolonged stratification into early autumn manifests in negative accumulation rates, along with pronounced signatures of compromised membranes, death-related protease activity, virus production, nutrient drawdown, and lipid markers indicative of nutrient stress. Positive accumulation renews during mixed layer deepening with transition into winter, concomitant with enhanced nutrient supply and lessened viral pressure.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Similar articles
-
Floats with bio-optical sensors reveal what processes trigger the North Atlantic bloom.Nat Commun. 2018 Jan 15;9(1):190. doi: 10.1038/s41467-017-02143-6. Nat Commun. 2018. PMID: 29335403 Free PMC article.
-
Latitudinal variation in virus-induced mortality of phytoplankton across the North Atlantic Ocean.ISME J. 2016 Feb;10(2):500-13. doi: 10.1038/ismej.2015.130. Epub 2015 Aug 11. ISME J. 2016. PMID: 26262815 Free PMC article.
-
Seasonality of North Atlantic phytoplankton from space: impact of environmental forcing on a changing phenology (1998-2012).Glob Chang Biol. 2014 Mar;20(3):698-712. doi: 10.1111/gcb.12352. Epub 2014 Jan 4. Glob Chang Biol. 2014. PMID: 23943398
-
Impact of solar ultraviolet radiation on marine phytoplankton of Patagonia, Argentina.Photochem Photobiol. 2005 Jul-Aug;81(4):807-18. doi: 10.1562/2005-03-02-RA-452. Photochem Photobiol. 2005. PMID: 15839753 Review.
-
The Impact of Submesoscale Physics on Primary Productivity of Plankton.Ann Rev Mar Sci. 2016;8:161-84. doi: 10.1146/annurev-marine-010814-015912. Epub 2015 Sep 21. Ann Rev Mar Sci. 2016. PMID: 26394203 Review.
Cited by
-
Virus infection of phytoplankton increases average molar mass and reduces hygroscopicity of aerosolized organic matter.Sci Rep. 2023 May 5;13(1):7361. doi: 10.1038/s41598-023-33818-4. Sci Rep. 2023. PMID: 37147322 Free PMC article.
-
Macroecological patterns of planktonic unicellular eukaryotes richness in the Southeast Pacific Ocean.Sci Rep. 2025 May 29;15(1):18833. doi: 10.1038/s41598-025-03220-3. Sci Rep. 2025. PMID: 40442169 Free PMC article.
-
Altered growth and death in dilution-based viral predation assays.PLoS One. 2023 Jul 7;18(7):e0288114. doi: 10.1371/journal.pone.0288114. eCollection 2023. PLoS One. 2023. PMID: 37418487 Free PMC article.
-
Global marine phytoplankton dynamics analysis with machine learning and reanalyzed remote sensing.PeerJ. 2024 May 8;12:e17361. doi: 10.7717/peerj.17361. eCollection 2024. PeerJ. 2024. PMID: 38737741 Free PMC article.
-
Marine phytoplankton downregulate core photosynthesis and carbon storage genes upon rapid mixed layer shallowing.ISME J. 2023 Jul;17(7):1074-1088. doi: 10.1038/s41396-023-01416-x. Epub 2023 May 8. ISME J. 2023. PMID: 37156837 Free PMC article.
References
-
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–240. - PubMed
-
- Valiela, I. Marine Ecological Processes (Springer, 1995).
-
- Sverdrup HU. On conditions for the vernal blooming of phytoplankton. ICES J. Mar. Sci. 1953 doi: 10.1093/icesjms/18.3.287. - DOI

