RNF19A-mediated ubiquitination of BARD1 prevents BRCA1/BARD1-dependent homologous recombination
- PMID: 34789768
- PMCID: PMC8599684
- DOI: 10.1038/s41467-021-27048-3
RNF19A-mediated ubiquitination of BARD1 prevents BRCA1/BARD1-dependent homologous recombination
Abstract
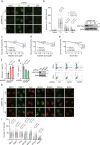
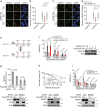
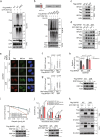
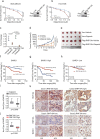
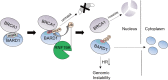
BRCA1-BARD1 heterodimers act in multiple steps during homologous recombination (HR) to ensure the prompt repair of DNA double strand breaks. Dysfunction of the BRCA1 pathway enhances the therapeutic efficiency of poly-(ADP-ribose) polymerase inhibitors (PARPi) in cancers, but the molecular mechanisms underlying this sensitization to PARPi are not fully understood. Here, we show that cancer cell sensitivity to PARPi is promoted by the ring between ring fingers (RBR) protein RNF19A. We demonstrate that RNF19A suppresses HR by ubiquitinating BARD1, which leads to dissociation of BRCA1-BARD1 complex and exposure of a nuclear export sequence in BARD1 that is otherwise masked by BRCA1, resulting in the export of BARD1 to the cytoplasm. We provide evidence that high RNF19A expression in breast cancer compromises HR and increases sensitivity to PARPi. We propose that RNF19A modulates the cancer cell response to PARPi by negatively regulating the BRCA1-BARD1 complex and inhibiting HR-mediated DNA repair.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
BARD1 reads H2A lysine 15 ubiquitination to direct homologous recombination.Nature. 2021 Aug;596(7872):433-437. doi: 10.1038/s41586-021-03776-w. Epub 2021 Jul 28. Nature. 2021. PMID: 34321663
-
SIRT2 promotes BRCA1-BARD1 heterodimerization through deacetylation.Cell Rep. 2021 Mar 30;34(13):108921. doi: 10.1016/j.celrep.2021.108921. Cell Rep. 2021. PMID: 33789098 Free PMC article.
-
Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.Nature. 2021 Aug;596(7872):438-443. doi: 10.1038/s41586-021-03716-8. Epub 2021 Jul 28. Nature. 2021. PMID: 34321665 Free PMC article.
-
The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication.Nat Rev Mol Cell Biol. 2020 May;21(5):284-299. doi: 10.1038/s41580-020-0218-z. Epub 2020 Feb 24. Nat Rev Mol Cell Biol. 2020. PMID: 32094664 Free PMC article. Review.
-
The BRCA1/BARD1 ubiquitin ligase and its substrates.Biochem J. 2021 Sep 30;478(18):3467-3483. doi: 10.1042/BCJ20200864. Biochem J. 2021. PMID: 34591954 Free PMC article. Review.
Cited by
-
Absence of Rnf126 causes male infertility with multiple morphological abnormalities of the sperm flagella.Cell Death Discov. 2025 May 23;11(1):251. doi: 10.1038/s41420-025-02432-w. Cell Death Discov. 2025. PMID: 40410177 Free PMC article.
-
PCAF-mediated acetylation regulates RAD51 dynamic localization on chromatin during HR repair.EMBO Rep. 2025 Aug;26(16):4100-4123. doi: 10.1038/s44319-025-00513-6. Epub 2025 Jul 15. EMBO Rep. 2025. PMID: 40664718
-
Crosstalk between SUMOylation and ubiquitylation controls DNA end resection by maintaining MRE11 homeostasis on chromatin.Nat Commun. 2022 Sep 1;13(1):5133. doi: 10.1038/s41467-022-32920-x. Nat Commun. 2022. PMID: 36050397 Free PMC article.
-
Chromatin Ubiquitination Guides DNA Double Strand Break Signaling and Repair.Front Cell Dev Biol. 2022 Jul 5;10:928113. doi: 10.3389/fcell.2022.928113. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35865631 Free PMC article. Review.
-
Homologous recombination mRNAs (RAD21, RAD50 and BARD1) have a potentially poor prognostic role in ERBB2-low bladder cancer patients.Sci Rep. 2023 Jul 20;13(1):11738. doi: 10.1038/s41598-023-38923-y. Sci Rep. 2023. PMID: 37474724 Free PMC article.
References
-
- Ochs F, et al. Stabilization of chromatin topology safeguards genome integrity. Nature. 2019;574:571–574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

