Schizophrenia is defined by cell-specific neuropathology and multiple neurodevelopmental mechanisms in patient-derived cerebral organoids
- PMID: 34789849
- PMCID: PMC9095467
- DOI: 10.1038/s41380-021-01316-6
Schizophrenia is defined by cell-specific neuropathology and multiple neurodevelopmental mechanisms in patient-derived cerebral organoids
Abstract
Due to an inability to ethically access developing human brain tissue as well as identify prospective cases, early-arising neurodevelopmental and cell-specific signatures of Schizophrenia (Scz) have remained unknown and thus undefined. To overcome these challenges, we utilized patient-derived induced pluripotent stem cells (iPSCs) to generate 3D cerebral organoids to model neuropathology of Scz during this critical period. We discovered that Scz organoids exhibited ventricular neuropathology resulting in altered progenitor survival and disrupted neurogenesis. This ultimately yielded fewer neurons within developing cortical fields of Scz organoids. Single-cell sequencing revealed that Scz progenitors were specifically depleted of neuronal programming factors leading to a remodeling of cell-lineages, altered differentiation trajectories, and distorted cortical cell-type diversity. While Scz organoids were similar in their macromolecular diversity to organoids generated from healthy controls (Ctrls), four GWAS factors (PTN, COMT, PLCL1, and PODXL) and peptide fragments belonging to the POU-domain transcription factor family (e.g., POU3F2/BRN2) were altered. This revealed that Scz organoids principally differed not in their proteomic diversity, but specifically in their total quantity of disease and neurodevelopmental factors at the molecular level. Single-cell sequencing subsequently identified cell-type specific alterations in neuronal programming factors as well as a developmental switch in neurotrophic growth factor expression, indicating that Scz neuropathology can be encoded on a cell-type-by-cell-type basis. Furthermore, single-cell sequencing also specifically replicated the depletion of BRN2 (POU3F2) and PTN in both Scz progenitors and neurons. Subsequently, in two mechanistic rescue experiments we identified that the transcription factor BRN2 and growth factor PTN operate as mechanistic substrates of neurogenesis and cellular survival, respectively, in Scz organoids. Collectively, our work suggests that multiple mechanisms of Scz exist in patient-derived organoids, and that these disparate mechanisms converge upon primordial brain developmental pathways such as neuronal differentiation, survival, and growth factor support, which may amalgamate to elevate intrinsic risk of Scz.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
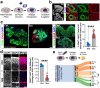

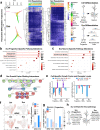
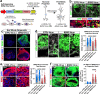
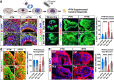
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

