Diversification of Escherichia albertii H-Antigens and Development of H-Genotyping PCR
- PMID: 34790177
- PMCID: PMC8591213
- DOI: 10.3389/fmicb.2021.737979
Diversification of Escherichia albertii H-Antigens and Development of H-Genotyping PCR
Abstract
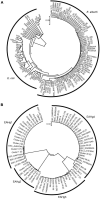
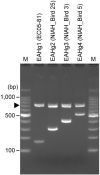
Escherichia albertii is a recently recognized human enteropathogen that is closely related to Escherichia coli. As E. albertii sometimes causes outbreaks of gastroenteritis, rapid strain typing systems, such as the O- and H-serotyping systems widely used for E. coli, will be useful for outbreak investigation and surveillance. Although an O-genotyping system has recently been developed, the diversity of E. albertii H-antigens (flagellins) encoded by fliC genes remains to be systematically investigated, and no H-serotyping or genotyping system is currently available. Here, we analyzed the fliC genes of 243 genome-sequenced E. albertii strains and identified 73 sequence types, which were grouped into four clearly distinguishable types designated E. albertii H-genotypes 1-4 (EAHg1-EAHg4). Although there was a clear sign of intraspecies transfer of fliC genes in E. albertii, none of the four E. albertii H-genotypes (EAHgs) were closely related to any of the 53 known E. coli H-antigens, indicating the absence or rare occurrence of interspecies transfer of fliC genes between the two species. Although the analysis of more E. albertii strains will be required to confirm the low level of variation in their fliC genes, this finding suggests that E. albertii may exist in limited natural hosts or environments and/or that the flagella of E. albertii may function in a limited stage(s) in their life cycle. Based on the fliC sequences of the four EAHgs, we developed a multiplex PCR-based H-genotyping system for E. albertii (EAH-genotyping PCR), which will be useful for epidemiological studies of E. albertii infections.
Keywords: Escherichia albertii; H-antigen; flagellin; fliC; genotyping.
Copyright © 2021 Nakae, Ooka, Murakami, Hara-Kudo, Imuta, Gotoh, Ogura, Hayashi, Okamoto and Nishi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Albert M. J., Faruque S. M., Ansaruzzaman M., Islam M. M., Haider K., Alam K., et al. (1992). Sharing of virulence-associated properties at the phenotypic and genetic levels between enteropathogenic Escherichia coli and Hafnia alvei. J. Med. Microbiol. 37 310–314. 10.1099/00222615-37-5-310 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

