Improved recognition of splice sites in A. thaliana by incorporating secondary structure information into sequence-derived features: a computational study
- PMID: 34790508
- PMCID: PMC8558126
- DOI: 10.1007/s13205-021-03036-8
Improved recognition of splice sites in A. thaliana by incorporating secondary structure information into sequence-derived features: a computational study
Abstract
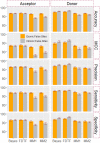
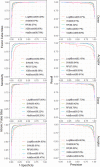
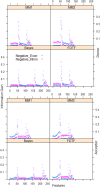
Identification of splice sites is an important aspect with regard to the prediction of gene structure. In most of the existing splice site prediction studies, machine learning algorithms coupled with sequence-derived features have been successfully employed for splice site recognition. However, the splice site identification by incorporating the secondary structure information is lacking, particularly in plant species. Thus, we made an attempt in this study to evaluate the performance of structural features on the splice site prediction accuracy in Arabidopsis thaliana. Prediction accuracies were evaluated with the sequence-derived features alone as well as by incorporating the structural features into the sequence-derived features, where support vector machine (SVM) was employed as prediction algorithm. Both short (40 base pairs) and long (105 base pairs) sequence datasets were considered for evaluation. After incorporating the secondary structure features, improvements in accuracies were observed only for the longer sequence dataset and the improvement was found to be higher with the sequence-derived features that accounted nucleotide dependencies. On the other hand, either a little or no improvement in accuracies was found for the short sequence dataset. The performance of SVM was further compared with that of LogitBoost, Random Forest (RF), AdaBoost and XGBoost machine learning methods. The prediction accuracies of SVM, AdaBoost and XGBoost were observed to be at par and higher than that of RF and LogitBoost algorithms. While prediction was performed by taking all the sequence-derived features along with the structural features, a little improvement in accuracies was found as compared to the combination of individual sequence-based features and structural features. To the best of our knowledge, this is the first attempt concerning the computational prediction of splice sites using machine learning methods by incorporating the secondary structure information into the sequence-derived features. All the source codes are available at https://github.com/meher861982/SSFeature.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-021-03036-8.
Keywords: Computational biology; Machine learning; Nucleotide dependencies; Secondary structure; Splice junction.
© King Abdulaziz City for Science and Technology 2021.
Conflict of interest statement
Conflict of interestThe author declares that there is no conflict of interest.
Figures


References
-
- Alfaro E, Gamez M, García N. adabag: an R package for classification with boosting and bagging. J Stat Softw. 2013;54:1–35. doi: 10.18637/jss.v054.i02. - DOI
-
- Bari ATM, Reaz M, Jeong B-S. Effective DNA encoding for splice site prediction using SVM. Match (mulheim an Der Ruhr, Germany) 2013;71:241–258.
-
- Breiman L. Random forests. Mach Learn. 2001 doi: 10.1023/A:1010933404324. - DOI
LinkOut - more resources
Full Text Sources

