Establishing and Validating an Aging-Related Prognostic Four-Gene Signature in Colon Adenocarcinoma
- PMID: 34790819
- PMCID: PMC8592697
- DOI: 10.1155/2021/4682589
Establishing and Validating an Aging-Related Prognostic Four-Gene Signature in Colon Adenocarcinoma
Retraction in
-
Retracted: Establishing and Validating an Aging-Related Prognostic Four-Gene Signature in Colon Adenocarcinoma.Biomed Res Int. 2024 Mar 20;2024:9817540. doi: 10.1155/2024/9817540. eCollection 2024. Biomed Res Int. 2024. PMID: 38550120 Free PMC article.
Abstract
Background: Aging is a process that biological changes accumulate with time and lead to increasing susceptibility to diseases like cancer. This study is aimed at establishing an aging-related prognostic signature in colon adenocarcinoma (COAD).
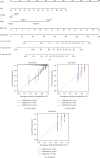
Methods: The transcriptome data and clinical variables of COAD patients were downloaded from TCGA database. The genes in GOBP_AGING gene set was used for prognostic evaluation by the univariate and multivariate Cox regression analyses. The model was presented by a nomogram and assessed by the Kaplan-Meier curves and calibration curves. The drug response and gene mutation were also performed to implicate the clinical significance. The GO and KEGG analyses were employed to unravel the potential functional mechanism.
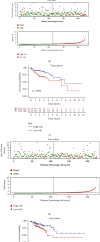
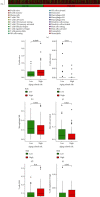
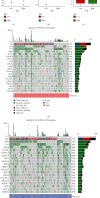
Results: The Gene Set Enrichment Analysis result indicates that GOBP_AGING pathway is significantly enriched in COAD samples. Four aging-related genes are finally used to construct the aging-related prognostic signature: FOXM1, PTH1R, KL, and CGAS. The COAD patients with high risk score have much shorter overall survival in both train cohort and test cohort. The nomogram is then assembled to predict 1-year, 3-year, and 5-year survival. Patients with high risk score have elevated infiltrating B cell naïve and attenuated cisplatin sensitivity. The mutation landscape shows that the TTN, FAT4, ZFHX4, APC, and OBSCN gene mutation are different between high risk score patients and low risk score patients. The differentially expressed genes between patients with high score and low score are enriched in B cell receptor signaling pathway.
Conclusion: We constructed an aging-related signature in COAD patients, which can predict oncological outcome and optimize therapeutic strategy.
Copyright © 2021 Lian Zheng et al.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

