Metatranscriptomics to characterize respiratory virome, microbiome, and host response directly from clinical samples
- PMID: 34790908
- PMCID: PMC8594859
- DOI: 10.1016/j.crmeth.2021.100091
Metatranscriptomics to characterize respiratory virome, microbiome, and host response directly from clinical samples
Abstract
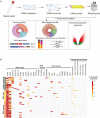
We developed a metatranscriptomics method that can simultaneously capture the respiratory virome, microbiome, and host response directly from low biomass samples. Using nasal swab samples, we capture RNA virome with sufficient sequencing depth required to assemble complete genomes. We find a surprisingly high frequency of respiratory syncytial virus (RSV) and coronavirus (CoV) in healthy children, and a high frequency of RSV-A and RSV-B co-detections in children with symptomatic RSV. In addition, we have identified commensal and pathogenic bacteria and fungi at the species level. Functional analysis revealed that H. influenzae was highly active in symptomatic RSV subjects. The host nasal transcriptome reveled upregulation of the innate immune system, anti-viral response and inflammasome pathway, and downregulation of fatty acid pathways in children with symptomatic RSV. Overall, we demonstrate that our method is broadly applicable to infer the transcriptome landscape of an infected system, surveil respiratory infections, and to sequence RNA viruses directly from clinical samples.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures





References
-
- Abbas A.A., Taylor L.J., Dothard M.I., Leiby J.S., Fitzgerald A.S., Khatib L.A., Collman R.G., Bushman F.D. Redondoviridae, a family of small, circular DNA viruses of the human oro-respiratory tract associated with periodontitis and critical illness. Cell Host Microbe. 2019;25:719–729 e714. - PMC - PubMed
-
- Antunes K.H., Fachi J.L., de Paula R., da Silva E.F., Pral L.P., Dos Santos A.A., Dias G.B.M., Vargas J.E., Puga R., Mayer F.Q., et al. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat. Commun. 2019;10:3273. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

