AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models
- PMID: 34791371
- PMCID: PMC8728224
- DOI: 10.1093/nar/gkab1061
AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models
Abstract
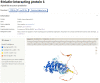
The AlphaFold Protein Structure Database (AlphaFold DB, https://alphafold.ebi.ac.uk) is an openly accessible, extensive database of high-accuracy protein-structure predictions. Powered by AlphaFold v2.0 of DeepMind, it has enabled an unprecedented expansion of the structural coverage of the known protein-sequence space. AlphaFold DB provides programmatic access to and interactive visualization of predicted atomic coordinates, per-residue and pairwise model-confidence estimates and predicted aligned errors. The initial release of AlphaFold DB contains over 360,000 predicted structures across 21 model-organism proteomes, which will soon be expanded to cover most of the (over 100 million) representative sequences from the UniRef90 data set.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Lee D., Redfern O., Orengo C.. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 2007; 8:995–1005. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

