An in situ sequencing approach maps PLASTOCHRON1 at the boundary between indeterminate and determinate cells
- PMID: 34791481
- PMCID: PMC8825424
- DOI: 10.1093/plphys/kiab533
An in situ sequencing approach maps PLASTOCHRON1 at the boundary between indeterminate and determinate cells
Abstract
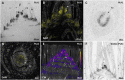
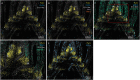
The plant shoot apex houses the shoot apical meristem, a highly organized and active stem-cell tissue where molecular signaling in discrete cells determines when and where leaves are initiated. We optimized a spatial transcriptomics approach, in situ sequencing (ISS), to colocalize the transcripts of 90 genes simultaneously on the same section of tissue from the maize (Zea mays) shoot apex. The RNA ISS technology reported expression profiles that were highly comparable with those obtained by in situ hybridizations (ISHs) and allowed the discrimination between tissue domains. Furthermore, the application of spatial transcriptomics to the shoot apex, which inherently comprised phytomers that are in gradual developmental stages, provided a spatiotemporal sequence of transcriptional events. We illustrate the power of the technology through PLASTOCHRON1 (PLA1), which was specifically expressed at the boundary between indeterminate and determinate cells and partially overlapped with ROUGH SHEATH1 and OUTER CELL LAYER4 transcripts. Also, in the inflorescence, PLA1 transcripts localized in cells subtending the lateral primordia or bordering the newly established meristematic region, suggesting a more general role of PLA1 in signaling between indeterminate and determinate cells during the formation of lateral organs. Spatial transcriptomics builds on RNA ISH, which assays relatively few transcripts at a time and provides a powerful complement to single-cell transcriptomics that inherently removes cells from their native spatial context. Further improvements in resolution and sensitivity will greatly advance research in plant developmental biology.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Besnard F, Refahi Y, Morin V, Marteaux B, Brunoud G, Chambrier P, Rozier F, Mirabet V, Legrand J, Lainé S, et al. (2014) Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505: 417–421 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

