Emergent SARS-CoV-2 variants: comparative replication dynamics and high sensitivity to thapsigargin
- PMID: 34793280
- PMCID: PMC8667886
- DOI: 10.1080/21505594.2021.2006960
Emergent SARS-CoV-2 variants: comparative replication dynamics and high sensitivity to thapsigargin
Abstract
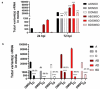
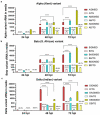
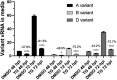
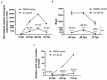
The struggle to control the COVID-19 pandemic is made challenging by the emergence of virulent SARS-CoV-2 variants. To gain insight into their replication dynamics, emergent Alpha (A), Beta (B) and Delta (D) SARS-CoV-2 variants were assessed for their infection performance in single variant- and co-infections. The effectiveness of thapsigargin (TG), a recently discovered broad-spectrum antiviral, against these variants was also examined. Of the 3 viruses, the D variant exhibited the highest replication rate and was most able to spread to in-contact cells; its replication rate at 24 h post-infection (hpi) based on progeny viral RNA production was over 4 times that of variant A and 9 times more than the B variant. In co-infections, the D variant boosted the replication of its co-infected partners at the expense of its own initial performance. Furthermore, co-infection with AD or AB combination conferred replication synergy where total progeny (RNA) output was greater than the sum of corresponding single-variant infections. All variants were highly sensitive to TG inhibition. A single pre-infection priming dose of TG effectively blocked all single-variant infections and every combination (AB, AD, BD variants) of co-infection at greater than 95% (relative to controls) at 72 hpi. Likewise, TG was effective in inhibiting each variant in active preexisting infection. In conclusion, against the current backdrop of the dominant D variant that could be further complicated by co-infection synergy with new variants, the growing list of viruses susceptible to TG, a promising host-centric antiviral, now includes a spectrum of contemporary SARS-CoV-2 viruses.
Keywords: Alpha; Beta; Delta; SARS-CoV-2; antiviral; co-infection; emergent variants; replication synergy; syncytia; thapsigargin.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. - PubMed
-
- Scudellari M. How the coronavirus infects cells — and why Delta is so dangerous. Nature. 2021;595(7869):640–644. - PubMed
-
- Thorne LG, Bouhaddou M, Reuschl AK, et al. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant. bioRxiv. 2021. DOI:10.1101/2021.06.06.446826 - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
