Nsp1 of SARS-CoV-2 stimulates host translation termination
- PMID: 34793288
- PMCID: PMC8782162
- DOI: 10.1080/15476286.2021.1999103
Nsp1 of SARS-CoV-2 stimulates host translation termination
Abstract
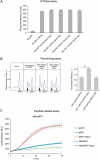
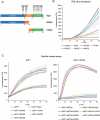
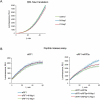
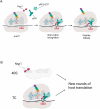
Nsp1 of SARS-CoV-2 regulates the translation of host and viral mRNAs in cells. Nsp1 inhibits host translation initiation by occluding the entry channel of the 40S ribosome subunit. The structural study of the Nsp1-ribosomal complexes reported post-termination 80S complex containing Nsp1, eRF1 and ABCE1. Considering the presence of Nsp1 in the post-termination 80S ribosomal complex, we hypothesized that Nsp1 may be involved in translation termination. Using a cell-free translation system and reconstituted in vitro translation system, we show that Nsp1 stimulates peptide release and formation of termination complexes. Detailed analysis of Nsp1 activity during translation termination stages reveals that Nsp1 facilitates stop codon recognition. We demonstrate that Nsp1 stimulation targets eRF1 and does not affect eRF3. Moreover, Nsp1 increases amount of the termination complexes at all three stop codons. The activity of Nsp1 in translation termination is provided by its N-terminal domain and the minimal required part of eRF1 is NM domain. We assume that the biological meaning of Nsp1 activity in translation termination is binding with the 80S ribosomes translating host mRNAs and remove them from the pool of the active ribosomes.
Keywords: Nsp1; SARS-CoV-2; eRF1; eRF3; ribosome.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
