Three-dimensional loop extrusion
- PMID: 34793758
- PMCID: PMC8715238
- DOI: 10.1016/j.bpj.2021.11.015
Three-dimensional loop extrusion
Abstract
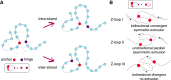
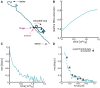
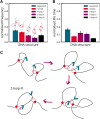
Loop extrusion convincingly describes how certain structural maintenance of chromosome (SMC) proteins mediate the formation of large DNA loops. Yet most of the existing computational models cannot reconcile recent in vitro observations showing that condensins can traverse each other, bypass large roadblocks, and perform steps longer than their own size. To fill this gap, we propose a three-dimensional (3D) "trans-grabbing" model for loop extrusion, which not only reproduces the experimental features of loop extrusion by one SMC complex but also predicts the formation of so-called Z-loops via the interaction of two or more SMCs extruding along the same DNA substrate. By performing molecular dynamics simulations of this model, we discover that the experimentally observed asymmetry in the different types of Z-loops is a natural consequence of the DNA tethering in vitro. Intriguingly, our model predicts this bias to disappear in the absence of tethering and a third type of Z-loop, which has not yet been identified in experiments, to appear. Our model naturally explains roadblock bypassing and the appearance of steps larger than the SMC size as a consequence of non-contiguous DNA grabbing. Finally, this study is the first, to our knowledge, to address how Z-loops and bypassing might occur in a way that is broadly consistent with existing cis-only 1D loop extrusion models.
Copyright © 2021 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat. Cell Biol. 2011;13:1170–1177. - PubMed
-
- Uhlmann F. SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell. Biol. 2016;17:399–412. - PubMed
-
- Hirano T. Condensin-based chromosome organization from bacteria to vertebrates. Cell. 2016;164:847–857. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

