Genome surveillance by HUSH-mediated silencing of intronless mobile elements
- PMID: 34794168
- PMCID: PMC8770142
- DOI: 10.1038/s41586-021-04228-1
Genome surveillance by HUSH-mediated silencing of intronless mobile elements
Abstract
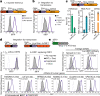
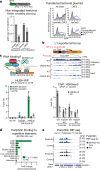
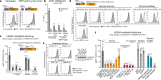
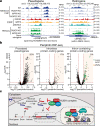
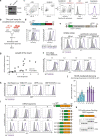
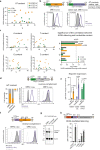
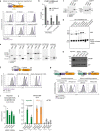
All life forms defend their genome against DNA invasion. Eukaryotic cells recognize incoming DNA and limit its transcription through repressive chromatin modifications. The human silencing hub (HUSH) complex transcriptionally represses long interspersed element-1 retrotransposons (L1s) and retroviruses through histone H3 lysine 9 trimethylation (H3K9me3)1-3. How HUSH recognizes and initiates silencing of these invading genetic elements is unknown. Here we show that HUSH is able to recognize and transcriptionally repress a broad range of long, intronless transgenes. Intron insertion into HUSH-repressed transgenes counteracts repression, even in the absence of intron splicing. HUSH binds transcripts from the target locus, prior to and independent of H3K9me3 deposition, and target transcription is essential for both initiation and propagation of HUSH-mediated H3K9me3. Genomic data reveal how HUSH binds and represses a subset of endogenous intronless genes generated through retrotransposition of cellular mRNAs. Thus intronless cDNA-the hallmark of reverse transcription-provides a versatile way to distinguish invading retroelements from host genes and enables HUSH to protect the genome from 'non-self' DNA, despite there being no previous exposure to the invading element. Our findings reveal the existence of a transcription-dependent genome-surveillance system and explain how it provides immediate protection against newly acquired elements while avoiding inappropriate repression of host genes.
© 2021. The Author(s).
Conflict of interest statement
S.M.C. is an employee of AstraZeneca. All other authors declare no competing interests.
Figures







References
-
- Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

