Optimization of non-coding regions for a non-modified mRNA COVID-19 vaccine
- PMID: 34794169
- PMCID: PMC8770133
- DOI: 10.1038/s41586-021-04231-6
Optimization of non-coding regions for a non-modified mRNA COVID-19 vaccine
Abstract
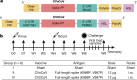
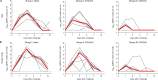
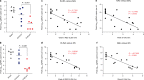
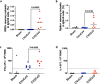
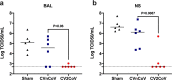
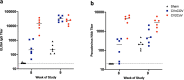
The CVnCoV (CureVac) mRNA vaccine for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was recently evaluated in a phase 2b/3 efficacy trial in humans1. CV2CoV is a second-generation mRNA vaccine containing non-modified nucleosides but with optimized non-coding regions and enhanced antigen expression. Here we report the results of a head-to-head comparison of the immunogenicity and protective efficacy of CVnCoV and CV2CoV in non-human primates. We immunized 18 cynomolgus macaques with two doses of 12 μg lipid nanoparticle-formulated CVnCoV or CV2CoV or with sham (n = 6 per group). Compared with CVnCoV, CV2CoV induced substantially higher titres of binding and neutralizing antibodies, memory B cell responses and T cell responses as well as more potent neutralizing antibody responses against SARS-CoV-2 variants, including the Delta variant. Moreover, CV2CoV was found to be comparably immunogenic to the BNT162b2 (Pfizer) vaccine in macaques. Although CVnCoV provided partial protection against SARS-CoV-2 challenge, CV2CoV afforded more robust protection with markedly lower viral loads in the upper and lower respiratory tracts. Binding and neutralizing antibody titres were correlated with protective efficacy. These data demonstrate that optimization of non-coding regions can greatly improve the immunogenicity and protective efficacy of a non-modified mRNA SARS-CoV-2 vaccine in non-human primates.
© 2021. The Author(s).
Conflict of interest statement
S.R., B.P., N.R. and S.O.M. are employees of CureVac AG, Tübingen, Germany, a publicly listed company developing mRNA-based vaccines and immunotherapeutics. Authors may hold shares in the company. S.R., B.P. and N.R. are inventors on several patents on mRNA vaccination and use thereof. The other authors declare no competing interests.
Figures





References
-
- Kremsner, P. G. et al. Efficacy and safety of the CVnCoV SARS-CoV-2 mRNA vaccine candidate: results from Herald, a phase 2b/3, randomised, observer-blinded, placebo-controlled clinical trial in ten countries in Europe and Latin America. Lancet Infect. Dis. 10.1016/S1473-3099(21)00677–0 (2021). - DOI - PMC - PubMed
-
- Roth, N. et al. CV2CoV, an enhanced mRNA-based SARS-CoV-2 vaccine candidate, supports higher protein expression and improved immunogenicity in rats. Preprint at 10.1101/2021.05.13.443734 (2021).
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

