Identification of proximal SUMO-dependent interactors using SUMO-ID
- PMID: 34795231
- PMCID: PMC8602451
- DOI: 10.1038/s41467-021-26807-6
Identification of proximal SUMO-dependent interactors using SUMO-ID
Abstract
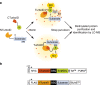
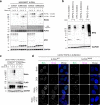
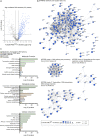
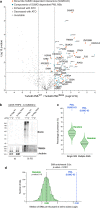
The fast dynamics and reversibility of posttranslational modifications by the ubiquitin family pose significant challenges for research. Here we present SUMO-ID, a technology that merges proximity biotinylation by TurboID and protein-fragment complementation to find SUMO-dependent interactors of proteins of interest. We develop an optimized split-TurboID version and show SUMO interaction-dependent labelling of proteins proximal to PML and RANGAP1. SUMO-dependent interactors of PML are involved in transcription, DNA damage, stress response and SUMO modification and are highly enriched in SUMO Interacting Motifs, but may only represent a subset of the total PML proximal proteome. Likewise, SUMO-ID also allow us to identify interactors of SUMOylated SALL1, a less characterized SUMO substrate. Furthermore, using TP53 as a substrate, we identify SUMO1, SUMO2 and Ubiquitin preferential interactors. Thus, SUMO-ID is a powerful tool that allows to study the consequences of SUMO-dependent interactions, and may further unravel the complexity of the ubiquitin code.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

