Defactinib inhibits PYK2 phosphorylation of IRF5 and reduces intestinal inflammation
- PMID: 34795257
- PMCID: PMC8602323
- DOI: 10.1038/s41467-021-27038-5
Defactinib inhibits PYK2 phosphorylation of IRF5 and reduces intestinal inflammation
Abstract
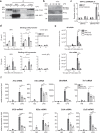
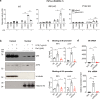
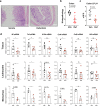
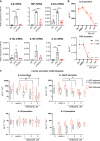
Interferon regulating factor 5 (IRF5) is a multifunctional regulator of immune responses, and has a key pathogenic function in gut inflammation, but how IRF5 is modulated is still unclear. Having performed a kinase inhibitor library screening in macrophages, here we identify protein-tyrosine kinase 2-beta (PTK2B/PYK2) as a putative IRF5 kinase. PYK2-deficient macrophages display impaired endogenous IRF5 activation, leading to reduction of inflammatory gene expression. Meanwhile, a PYK2 inhibitor, defactinib, has a similar effect on IRF5 activation in vitro, and induces a transcriptomic signature in macrophages similar to that caused by IRF5 deficiency. Finally, defactinib reduces pro-inflammatory cytokines in human colon biopsies from patients with ulcerative colitis, as well as in a mouse colitis model. Our results thus implicate a function of PYK2 in regulating the inflammatory response in the gut via the IRF5 innate sensing pathway, thereby opening opportunities for related therapeutic interventions for inflammatory bowel diseases and other inflammatory conditions.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

