Inflammasome-mediated GSDMD activation facilitates escape of Candida albicans from macrophages
- PMID: 34795266
- PMCID: PMC8602704
- DOI: 10.1038/s41467-021-27034-9
Inflammasome-mediated GSDMD activation facilitates escape of Candida albicans from macrophages
Abstract
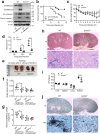
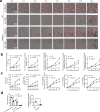
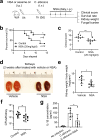
Candida albicans is the most common cause of fungal sepsis. Inhibition of inflammasome activity confers resistance to polymicrobial and LPS-induced sepsis; however, inflammasome signaling appears to protect against C. albicans infection, so inflammasome inhibitors are not clinically useful for candidiasis. Here we show disruption of GSDMD, a known inflammasome target and key pyroptotic cell death mediator, paradoxically alleviates candidiasis, improving outcomes and survival of Candida-infected mice. Mechanistically, C. albicans hijacked the canonical inflammasome-GSDMD axis-mediated pyroptosis to promote their escape from macrophages, deploying hyphae and candidalysin, a pore-forming toxin expressed by hyphae. GSDMD inhibition alleviated candidiasis by preventing C. albicans escape from macrophages while maintaining inflammasome-dependent but GSDMD-independent IL-1β production for anti-fungal host defenses. This study demonstrates key functions for GSDMD in Candida's escape from host immunity in vitro and in vivo and suggests that GSDMD may be a potential therapeutic target in C. albicans-induced sepsis.
© 2021. The Author(s).
Conflict of interest statement
All authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

