Intercellular nanotubes mediate mitochondrial trafficking between cancer and immune cells
- PMID: 34795441
- PMCID: PMC10071558
- DOI: 10.1038/s41565-021-01000-4
Intercellular nanotubes mediate mitochondrial trafficking between cancer and immune cells
Abstract
Cancer progresses by evading the immune system. Elucidating diverse immune evasion strategies is a critical step in the search for next-generation immunotherapies for cancer. Here we report that cancer cells can hijack the mitochondria from immune cells via physical nanotubes. Mitochondria are essential for metabolism and activation of immune cells. By using field-emission scanning electron microscopy, fluorophore-tagged mitochondrial transfer tracing and metabolic quantification, we demonstrate that the nanotube-mediated transfer of mitochondria from immune cells to cancer cells metabolically empowers the cancer cells and depletes the immune cells. Inhibiting the nanotube assembly machinery significantly reduced mitochondrial transfer and prevented the depletion of immune cells. Combining a farnesyltransferase and geranylgeranyltransferase 1 inhibitor, namely, L-778123, which partially inhibited nanotube formation and mitochondrial transfer, with a programmed cell death protein 1 immune checkpoint inhibitor improved the antitumour outcomes in an aggressive immunocompetent breast cancer model. Nanotube-mediated mitochondrial hijacking can emerge as a novel target for developing next-generation immunotherapy agents for cancer.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures
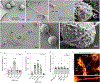
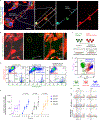



Comment in
-
Cancer cells hijack T-cell mitochondria.Nat Nanotechnol. 2022 Jan;17(1):3-4. doi: 10.1038/s41565-021-01006-y. Nat Nanotechnol. 2022. PMID: 34795442 No abstract available.
References
-
- Sharma P & Allison JP Dissecting the mechanisms of immune checkpoint therapy. Nat. Rev. Immunol 20, 75–76 (2020). - PubMed
-
- Wolchok J Putting the immunologic brakes on cancer. Cell 175, 1452–1454 (2018). - PubMed
-
- Wei SC, Duffy CR & Allison JP Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8, 1069–1086 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

